Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.plotting.displays.YZProjector¶
- class nilearn.plotting.displays.YZProjector(cut_coords, axes=None, black_bg=False, brain_color=(0.5, 0.5, 0.5), **kwargs)[source]¶
The
YZProjectorclass enables to combine coronal and axial views on the same figure through 2D projections.This visualization mode can be activated by setting
display_mode='yz':from nilearn.datasets import load_mni152_template from nilearn.plotting import plot_glass_brain img = load_mni152_template() # display is an instance of the YZProjector class display = plot_glass_brain(img, display_mode="yz")
- Parameters:
- cut_coordssequence of
floatorint The world coordinates of the point where the cut is performed. Not used for projectors.
- axes
matplotlib.axes.Axes, or 4tupleoffloat: (xmin, ymin, width, height), default=None The axes, or the coordinates, in matplotlib figure space, of the axes used to display the plot. If None, the complete figure is used.
- black_bg
bool, or “auto”, optional If True, the background of the image is set to be black. If you wish to save figures with a black background, you will need to pass facecolor=”k”, edgecolor=”k” to
matplotlib.pyplot.savefig.- brain_color
tuple, default=(0.5, 0.5, 0.5) The brain color to use as the background color (e.g., for transparent colorbars).
- kwargs
dict Extra keyword arguments are passed to
GlassBrainAxesused for plotting in each direction.
- cut_coordssequence of
- Attributes:
- cut_coords(None, None)
The world coordinates of the point where the cut is performed.
- axes
dictofGlassBrainAxes The axes used for plotting in each direction (‘y’ and ‘z’ here).
- frame_axes
Axes The axes framing the whole set of views.
- rect4-
tuple Position of axes within the figure.
See also
nilearn.plotting.displays.XZProjectorSagittal + Axial views
nilearn.plotting.displays.YXProjectorCoronal + Sagittal views
- add_contours(img, threshold=1e-06, filled=False, **kwargs)[source]¶
Contour a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. Provides image to plot.
- threshold
intorfloatorNone, default=1e-6 Threshold to apply:
If
Noneis given, the maps are not thresholded.- If number is given, it must be non-negative. The specified
value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent.
- filled
bool, default=False If
filled=True, contours are displayed with color fillings.- kwargs
dict Extra keyword arguments are passed to function
contour, or functioncontourf. Useful, arguments are typical “levels”, which is a list of values to use for plotting a contour or contour fillings (iffilled=True), and “colors”, which is one color or a list of colors for these contours.
- Raises:
- ValueError
if the specified threshold is a negative number
Notes
If colors are not specified, default coloring choices (from matplotlib) for contours and contour_fillings can be different.
- add_edges(img, color='r')[source]¶
Plot the edges of a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. The 3D map to be plotted. If it is a masked array, only the non-masked part will be plotted.
- colormatplotlib color:
stror (r, g, b) value, default=’r’ The color used to display the edge map.
- add_graph(adjacency_matrix, node_coords, node_color='auto', node_size=50, edge_cmap='RdBu_r', edge_vmin=None, edge_vmax=None, edge_threshold=None, edge_kwargs=None, node_kwargs=None, colorbar=False)[source]¶
Plot undirected graph on each of the axes.
- Parameters:
- adjacency_matrix
numpy.ndarrayof shape(n, n) Represents the edges strengths of the graph. The matrix can be symmetric which will result in an undirected graph, or not symmetric which will result in a directed graph.
- node_coords
numpy.ndarrayof shape(n, 3) 3D coordinates of the graph nodes in world space.
- node_colorcolor or sequence of colors, default=’auto’
Color(s) of the nodes.
- node_sizescalar or array_like, default=50
Size(s) of the nodes in points^2.
- edge_cmap
Colormap, default=”RdBu_r” Colormap used for representing the strength of the edges.
- edge_vmin, edge_vmax
floator None, default=None If not
None, either or both of these values will be used to as the minimum and maximum values to color edges.If
Noneare supplied, the maximum absolute value within the given threshold will be used as minimum (multiplied by -1) and maximum coloring levels.
- edge_threshold
strorintorfloator None, default=None If it is a number only the edges with a value greater than
edge_thresholdwill be shown.If it is a string it must finish with a percent sign, e.g. “25.3%”, and only the edges with a abs(value) above the given percentile will be shown.
- edge_kwargs
dict, default=None Will be passed as kwargs for each edge
Line2D.- node_kwargs
dict, default=None Will be passed as kwargs to the function
scatterwhich plots all the nodes at one.- colorbar
bool, optional If True, display a colorbar next to the plots. Default=False.
- adjacency_matrix
- add_markers(marker_coords, marker_color='r', marker_size=30, **kwargs)[source]¶
Add markers to the plot.
- Parameters:
- marker_coords
ndarrayof shape(n_markers, 3) Coordinates of the markers to plot. For each slice, only markers that are 2 millimeters away from the slice are plotted.
- marker_colorpyplot compatible color or
listof shape(n_markers,), default=’r’ List of colors for each marker that can be string or matplotlib colors.
- marker_size
floatorlistoffloatof shape(n_markers,), default=30 Size in pixel for each marker.
- kwargs
dict Extra keyword arguments are passed to
matplotlib.pyplot.scatter.
- marker_coords
- add_overlay(img, threshold=1e-06, colorbar=False, cbar_tick_format='%.2g', cbar_vmin=None, cbar_vmax=None, transparency=None, transparency_range=None, **kwargs)[source]¶
Plot a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. If it is a masked array, only the non-masked part will be plotted.
- threshold
intorfloatorNone, default=1e-6 Threshold to apply:
If
Noneis given, the maps are not thresholded.- If number is given, it must be non-negative. The specified
value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent.
- colorbar
bool, optional If True, display a colorbar next to the plots. Default=False.
- cbar_tick_formatstr, default=”%.2g” (scientific notation)
Controls how to format the tick labels of the colorbar. Ex: use “%i” to display as integers.
- cbar_vmin
float, default=None Minimal value for the colorbar. If None, the minimal value is computed based on the data.
- cbar_vmax
float, default=None Maximal value for the colorbar. If None, the maximal value is computed based on the data.
- transparency
floatbetween 0 and 1, or a Niimg-Like object, or None, default = None Value to be passed as alpha value to
imshow. ifNoneis passed, it will be set to 1. If an image is passed, voxel-wise alpha blending will be applied, by relying on the absolute value oftransparencyat each voxel.Added in Nilearn 0.12.0.
- transparency_range
tupleorlistof 2 non-negative numbers, or None, default = None When an image is passed to
transparency, this determines the range of values in the image to use for transparency (alpha blending). For example withtransparency_range = [1.96, 3], any voxel / vertex (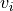 ):
):with a value between between -1.96 and 1.96, would be fully transparent (alpha = 0),
with a value less than -3 or greater than 3, would be fully opaque (alpha = 1),
with a value in the intervals
[-3.0, -1.96]or[1.96, 3.0], would have an alpha_i value scaled linearly between 0 and 1 :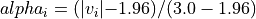 .
.
This parameter will be ignored unless an image is passed as
transparency. The first number must be greater than 0 and less than the second one. ifNoneis passed, this will be set to[0, max(abs(transparency))].Added in Nilearn 0.12.0.
- kwargs
dict Extra keyword arguments are passed to function
imshow.
- Raises:
- ValueError
if the specified threshold is a negative number
- annotate(left_right=True, positions=True, scalebar=False, size=12, scale_size=5.0, scale_units='cm', scale_loc=4, decimals=0, **kwargs)[source]¶
Add annotations to the plot.
- Parameters:
- left_right
bool, default=True If
True, annotations indicating which side is left and which side is right are drawn.- positions
bool, default=True If
True, annotations indicating the positions of the cuts are drawn.- scalebar
bool, default=False If
True, cuts are annotated with a reference scale bar. For finer control of the scale bar, please check out thedraw_scale_barmethod on the axes in “axes” attribute of this object.- size
int, default=12 The size of the text used.
- scale_size
intorfloat, default=5.0 The length of the scalebar, in units of
scale_units.- scale_units{‘cm’, ‘mm’}, default=’cm’
The units for the
scalebar.- scale_loc
int, default=4 The positioning for the scalebar. Valid location codes are:
1: “upper right”
2: “upper left”
3: “lower left”
4: “lower right”
5: “right”
6: “center left”
7: “center right”
8: “lower center”
9: “upper center”
10: “center”
- decimals
int, default=0 Number of decimal places on slice position annotation. If zero, the slice position is integer without decimal point.
- kwargs
dict Extra keyword arguments are passed to matplotlib’s text function.
- left_right
- property black_bg¶
Return black background.
- property brain_color¶
Return brain color.
- draw_cross(cut_coords=None, **kwargs)[source]¶
Draw a crossbar on the plot to show where the cut is performed.
Not implemented for this projector.
It does not make sense to draw crosses for the position of the cuts since we are taking the max along one axis.
- classmethod find_cut_coords(img=None, threshold=None, cut_coords=None)[source]¶
Return a tuple of None of length corresponding to the number of directions of this projector.
Specified parameters are not used.
- Parameters:
- img3D
Nifti1Image, default=None The brain image.
- threshold
intorfloator None, default=None Threshold to apply.
- cut_coordssequence of
floatorint, or None, default=None The world coordinates of the point where the cut is performed.
- img3D
- Returns:
- cut_coords
tupleof None tuple of None of length corresponding to the number of directions of this projector.
- cut_coords
- classmethod init_with_figure(img, threshold=None, cut_coords=None, figure=None, axes=None, black_bg=False, leave_space=False, colorbar=False, brain_color=(0.5, 0.5, 0.5), **kwargs)[source]¶
Initialize the slicer with an image.
- Parameters:
- imgNiimg-like object
- threshold
intorfloat, None, or ‘auto’, optional If None is given, the image is not thresholded. If number is given, it must be non-negative. The specified value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent. If “auto” is given, the threshold is determined based on the score obtained using percentile value “80%” on the absolute value of the image data.
- cut_coordsNone, allowed types depend on the
display_mode, optional The world coordinates of the point where the cut is performed.
If
display_modeis'ortho'or'tiled', this must be a 3-sequence offloatorint:(x, y, z).If
display_modeis'xz','yz'or'yx', this must be a 2-sequence offloatorint:(x, z),(y, z)or(x, y).If
display_modeis"x","y", or"z", this can be:If
display_modeis'mosaic', this can be:an
intin which case it specifies the number of cuts to perform in each direction"x","y","z".a 3-sequence of
floatorintin which case it specifies the number of cuts to perform in each direction"x","y","z"separately.dict<str: 1Dndarray> in which case keys are the directions (‘x’, ‘y’, ‘z’) and the values are sequences holding the cut coordinates.
If
Noneis given, the cuts are calculated automatically.
- figure
matplotlib.figure.Figure Figure to be used for plots.
- axes
matplotlib.axes.Axes, or 4tupleoffloat: (xmin, ymin, width, height), default=None The axes, or the coordinates, in matplotlib figure space, of the axes used to display the plot. If None, the complete figure is used. default=None The axes that will be subdivided in 3.
- black_bg
bool, or “auto”, optional If True, the background of the image is set to be black. If you wish to save figures with a black background, you will need to pass facecolor=”k”, edgecolor=”k” to
matplotlib.pyplot.savefig. default=False- leave_space
bool, default=False If
True, leave space between the plots.- colorbar
bool, optional If True, display a colorbar next to the plots. Default=False.
- brain_color
tuple, default=(0.5, 0.5, 0.5) The brain color to use as the background color (e.g., for transparent colorbars).
- kwargs
dict Extra keyword arguments are passed to
CutAxesused for plotting in each direction.
- Raises:
- ValueError
if the specified threshold is a negative number
- savefig(filename, dpi=None, **kwargs)[source]¶
Save the figure to a file.
- Parameters:
- filename
str The file name to save to. Its extension determines the file type, typically ‘.png’, ‘.svg’ or ‘.pdf’.
- dpiNone or scalar, default=None
The resolution in dots per inch.
- kwargs
dict Extra keyword arguments are passed to
matplotlib.pyplot.savefig.
- filename
- title(text, x=0.01, y=0.99, size=15, color=None, bgcolor=None, alpha=1, **kwargs)[source]¶
Write a title to the view.
- Parameters:
- text
str The text of the title.
- x
float, default=0.01 The horizontal position of the title on the frame in fraction of the frame width.
- y
float, default=0.99 The vertical position of the title on the frame in fraction of the frame height.
- size
int, default=15 The size of the title text.
- colormatplotlib color specifier, default=None
The color of the font of the title.
- bgcolormatplotlib color specifier, default=None
The color of the background of the title.
- alpha
float, default=1 The alpha value for the background.
- kwargs
dict Extra keyword arguments are passed to matplotlib’s text function.
- text