Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Plotting tools in nilearn¶
Nilearn comes with a set of plotting functions for easy visualization of Nifti-like images such as statistical maps mapped onto anatomical images or onto glass brain representation, anatomical images, functional/EPI images, region specific mask images.
See Plotting brain images for more details.
We will first retrieve data from nilearn provided (general-purpose) datasets.
from nilearn import datasets
# haxby dataset to have EPI images and masks
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print(
f"First subject anatomical nifti image (3D) is at: {haxby_dataset.anat[0]}"
)
print(
f"First subject functional nifti image (4D) is at: {haxby_dataset.func[0]}"
)
haxby_anat_filename = haxby_dataset.anat[0]
haxby_mask_filename = haxby_dataset.mask_vt[0]
haxby_func_filename = haxby_dataset.func[0]
# one motor activation map
stat_img = datasets.load_sample_motor_activation_image()
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
First subject anatomical nifti image (3D) is at: /home/runner/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Nilearn plotting functions¶
Plotting statistical maps: plot_stat_map¶
from nilearn import plotting
Visualizing t-map image on EPI template with manual positioning of coordinates using cut_coords given as a list
plotting.plot_stat_map(
stat_img, threshold=3, title="plot_stat_map", cut_coords=[36, -27, 66]
)
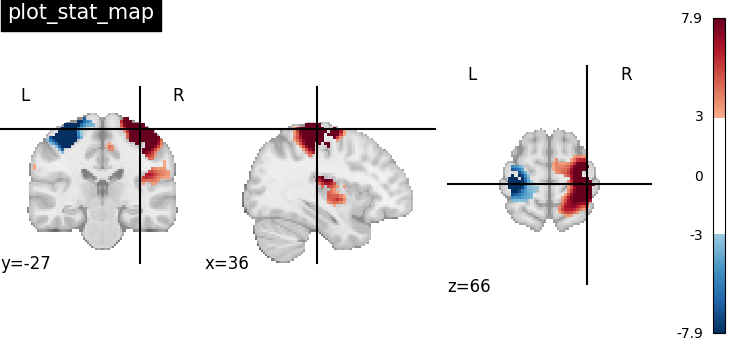
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f321d7100>
It’s also possible to visualize volumes in a LR-flipped “radiological” view Just set radiological=True
plotting.plot_stat_map(
stat_img,
threshold=3,
title="plot_stat_map",
cut_coords=[36, -27, 66],
radiological=True,
)
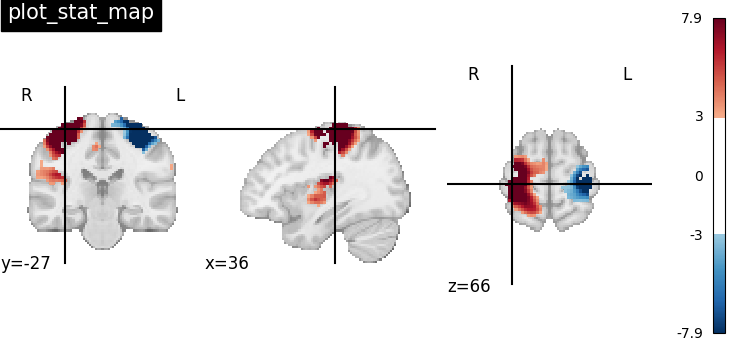
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f42116ad0>
Making interactive visualizations: view_img¶
An alternative to plot_stat_map is to use
view_img that gives more interactive
visualizations in a web browser. See Interactive visualization of statistical map slices
for more details.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.
uncomment this to open the plot in a web browser: view.open_in_browser()
It’s also possible to visualize volumes in a LR-flipped “radiological” view Just set radiological=True
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.
uncomment this to open the plot in a web browser: view_radio.open_in_browser()
Plotting statistical maps in a glass brain: plot_glass_brain¶
Now, the t-map image is mapped on glass brain representation where glass brain is always a fixed background template
plotting.plot_glass_brain(stat_img, title="plot_glass_brain", threshold=3)
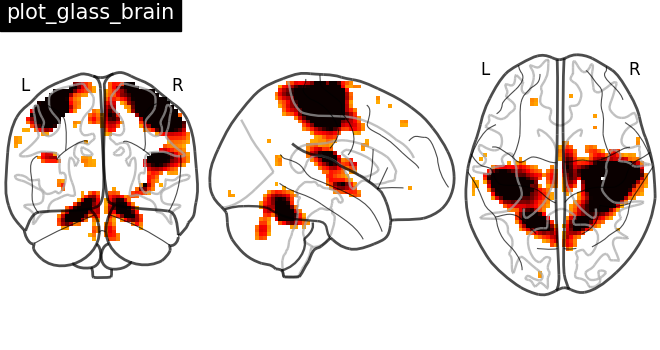
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f6f1dd172b0>
Plotting anatomical images: plot_anat¶
Visualizing anatomical image of haxby dataset
plotting.plot_anat(haxby_anat_filename, title="plot_anat")
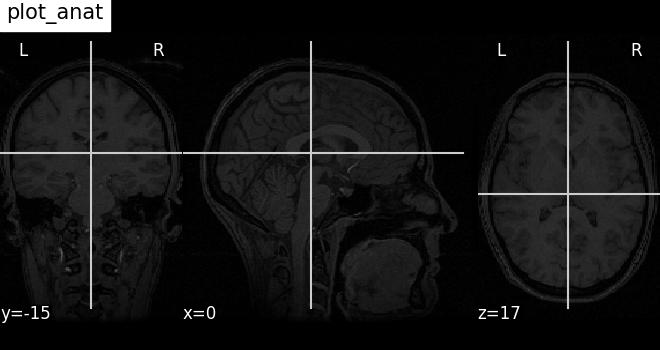
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6ed6a7b610>
Plotting ROIs (here the mask): plot_roi¶
Visualizing ventral temporal region image from haxby dataset overlaid on subject specific anatomical image with coordinates positioned automatically on region of interest (roi)
plotting.plot_roi(
haxby_mask_filename, bg_img=haxby_anat_filename, title="plot_roi"
)
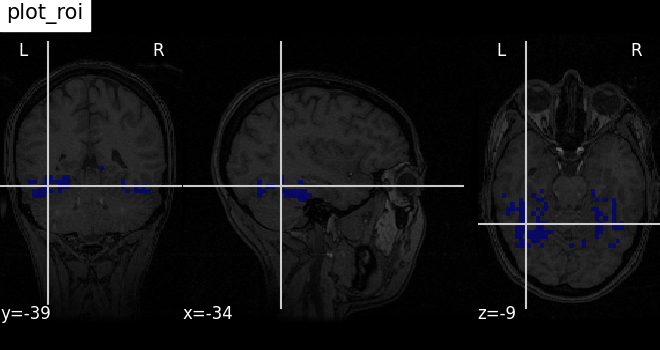
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6ef6d1e6b0>
Plotting EPI image: plot_epi¶
# Import image processing tool
from nilearn import image
# Compute the voxel_wise mean of functional images across time.
# Basically reducing the functional image from 4D to 3D
mean_haxby_img = image.mean_img(haxby_func_filename)
# Visualizing mean image (3D)
plotting.plot_epi(mean_haxby_img, title="plot_epi")
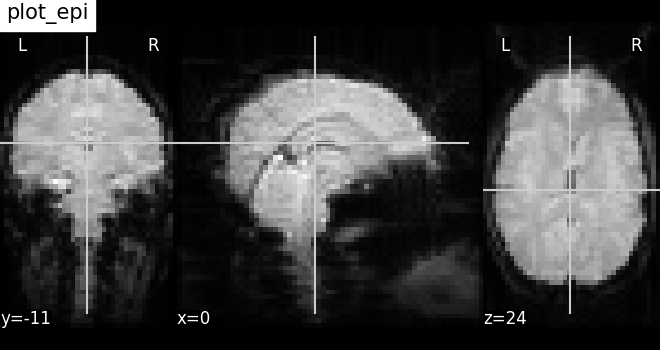
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f4162d390>
A call to plotting.show is needed to display the plots when running in script mode (ie outside IPython)
Thresholding plots¶
Using threshold value alongside with vmin and vmax parameters
enable us to mask certain values in the image.
Plotting without threshold¶
plotting.plot_stat_map(
stat_img,
display_mode="ortho",
cut_coords=[36, -27, 60],
title="No plotting threshold",
)
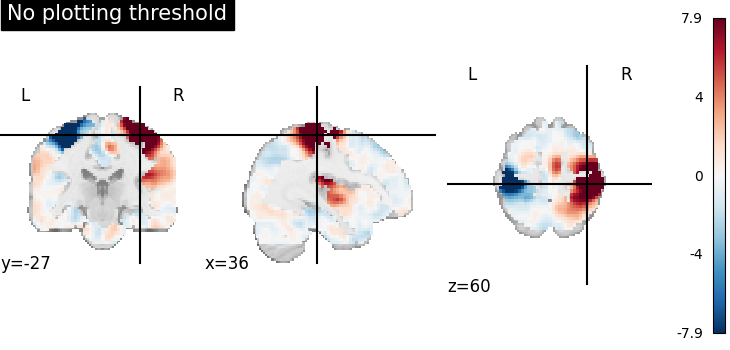
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f419d8b50>
Plotting threshold set to 1¶
When plotting threshold is set to 1, the values between -1 and 1 are masked in the plot.
plotting.plot_stat_map(
stat_img,
threshold=1,
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1",
)
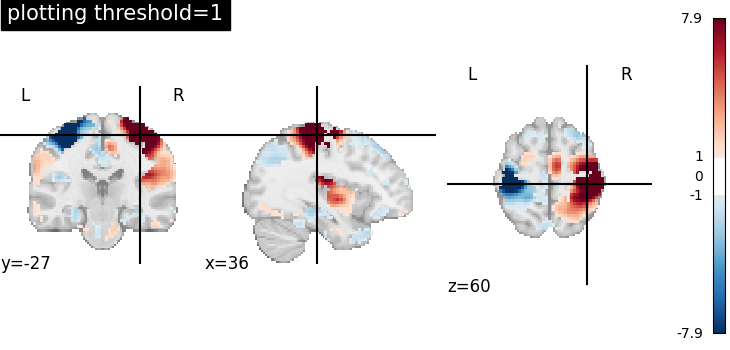
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f3f227f10>
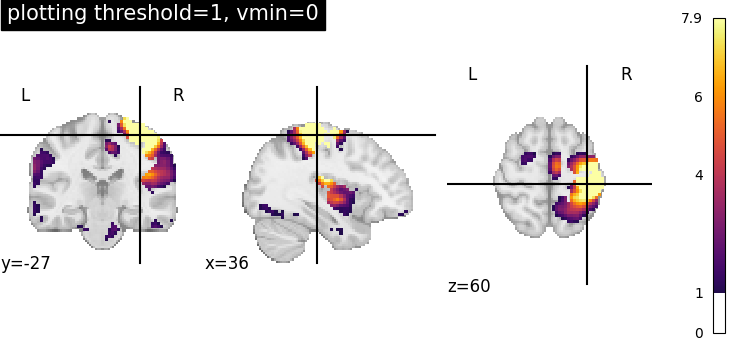
Plotting threshold set to 1 with vmin=0¶
Setting vmin=0, it is possible to plot only positive image values.
plotting.plot_stat_map(
stat_img,
threshold=1,
cmap="inferno",
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1, vmin=0",
vmin=0,
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f3fbc85e0>
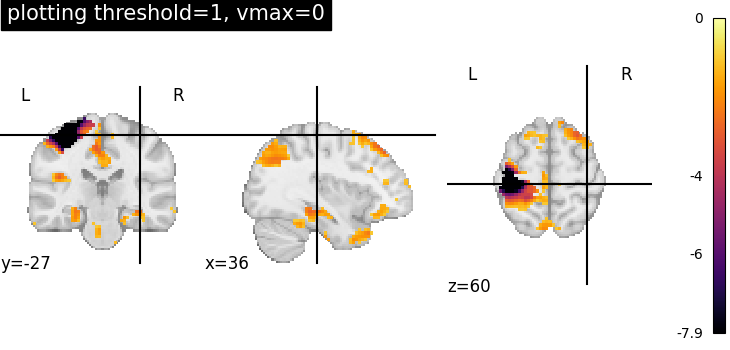
Plotting threshold set to 1 with vmax=0¶
Setting vmax=0, it is possible to plot only negative image values.
plotting.plot_stat_map(
stat_img,
threshold=1,
cmap="inferno",
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1, vmax=0",
vmax=0,
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f41abd510>
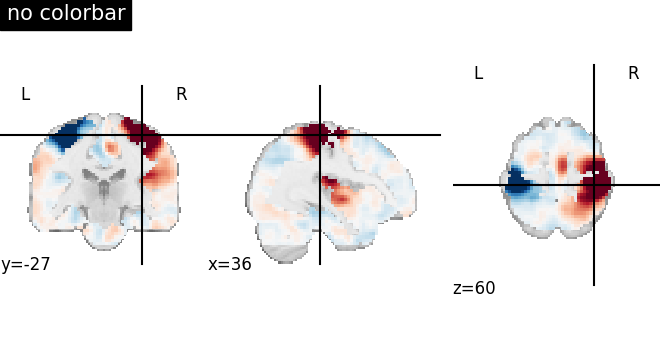
Visualizing without a colorbar on the right side¶
The argument colorbar should be set to False to show plots without
a colorbar on the right side.
plotting.plot_stat_map(
stat_img,
display_mode="ortho",
cut_coords=[36, -27, 60],
colorbar=False,
title="no colorbar",
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f3feeae00>
Total running time of the script: (0 minutes 13.521 seconds)
Estimated memory usage: 1014 MB