Note
Go to the end to download the full example code or to run this example in your browser via Binder.
A introduction tutorial to fMRI decoding¶
Here is a simple tutorial on decoding with nilearn. It reproduces the Haxby et al.[1] study on a face vs cat discrimination task in a mask of the ventral stream.
This tutorial is meant as an introduction to the various steps of a decoding
analysis using Nilearn meta-estimator: Decoder
It is not a minimalistic example, as it strives to be didactic. It is not meant to be copied to analyze new data: many of the steps are unnecessary.
import warnings
warnings.filterwarnings(
"ignore", message="The provided image has no sform in its header."
)
Retrieve and load the fMRI data from the Haxby study¶
First download the data¶
The fetch_haxby function will download the
Haxby dataset if not present on the disk, in the nilearn data directory.
It can take a while to download about 310 Mo of data from the Internet.
from nilearn import datasets
# By default 2nd subject will be fetched
haxby_dataset = datasets.fetch_haxby()
# 'func' is a list of filenames: one for each subject
fmri_filename = haxby_dataset.func[0]
# print basic information on the dataset
print(f"First subject functional nifti images (4D) are at: {fmri_filename}")
[fetch_haxby] Dataset created in /home/runner/nilearn_data/haxby2001
[fetch_haxby] Downloading data from
https://www.nitrc.org/frs/download.php/7868/mask.nii.gz ...
[fetch_haxby] ...done. (0 seconds, 0 min)
[fetch_haxby] Downloading data from
http://data.pymvpa.org/datasets/haxby2001/MD5SUMS ...
[fetch_haxby] ...done. (0 seconds, 0 min)
[fetch_haxby] Downloading data from
http://data.pymvpa.org/datasets/haxby2001/subj2-2010.01.14.tar.gz ...
[fetch_haxby] Downloaded 29679616 of 291168628 bytes (10.2%%, 8.8s remaining)
[fetch_haxby] Downloaded 85884928 of 291168628 bytes (29.5%%, 4.8s remaining)
[fetch_haxby] Downloaded 144105472 of 291168628 bytes (49.5%%, 3.1s
remaining)
[fetch_haxby] Downloaded 196829184 of 291168628 bytes (67.6%%, 1.9s
remaining)
[fetch_haxby] Downloaded 252944384 of 291168628 bytes (86.9%%, 0.8s
remaining)
[fetch_haxby] ...done. (6 seconds, 0 min)
[fetch_haxby] Extracting data from
/home/runner/nilearn_data/haxby2001/9cabe068089e791ef0c5fe930fc20e30/subj2-2010.
01.14.tar.gz...
[fetch_haxby] .. done.
First subject functional nifti images (4D) are at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
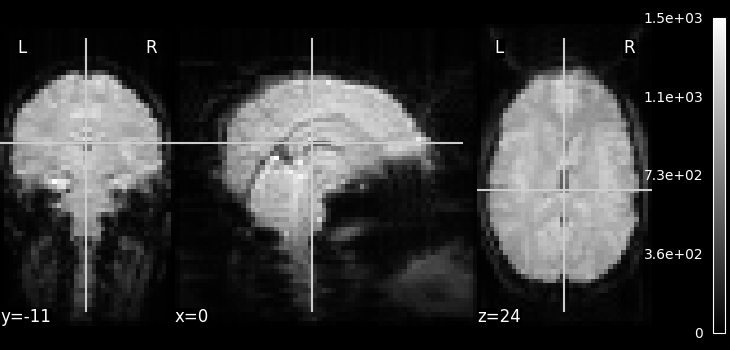
Visualizing the fMRI volume¶
One way to visualize a fMRI volume is
using plot_epi.
We will visualize the previously fetched fMRI
data from Haxby dataset.
Because fMRI data are 4D
(they consist of many 3D EPI images),
we cannot plot them directly using plot_epi
(which accepts just 3D input).
Here we are using mean_img to
extract a single 3D EPI image from the fMRI data.
Feature extraction: from fMRI volumes to a data matrix¶
These are some really lovely images, but for machine learning
we need matrices to work with the actual data. Fortunately, the
Decoder object we will use later on can
automatically transform Nifti images into matrices.
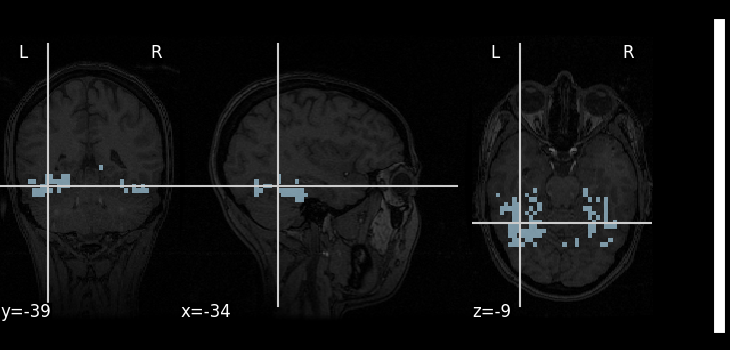
All we have to do for now is define a mask filename.
A mask of the Ventral Temporal (VT) cortex coming from the Haxby study is available:
mask_filename = haxby_dataset.mask_vt[0]
# Let's visualize it, using the subject's anatomical image as a
# background
plot_roi(mask_filename, bg_img=haxby_dataset.anat[0], cmap="Paired")
show()
Load the behavioral labels¶
Now that the brain images are converted to a data matrix, we can apply machine-learning to them, for instance to predict the task that the subject was doing. The behavioral labels are stored in a CSV file, separated by spaces.
We use pandas to load them in an array.
import pandas as pd
# Load behavioral information
behavioral = pd.read_csv(haxby_dataset.session_target[0], delimiter=" ")
print(behavioral)
labels chunks
0 rest 0
1 rest 0
2 rest 0
3 rest 0
4 rest 0
... ... ...
1447 rest 11
1448 rest 11
1449 rest 11
1450 rest 11
1451 rest 11
[1452 rows x 2 columns]
The task was a visual-recognition task, and the labels denote the experimental condition: the type of object that was presented to the subject. This is what we are going to try to predict.
conditions = behavioral["labels"]
print(conditions)
0 rest
1 rest
2 rest
3 rest
4 rest
...
1447 rest
1448 rest
1449 rest
1450 rest
1451 rest
Name: labels, Length: 1452, dtype: object
Restrict the analysis to cats and faces¶
As we can see from the targets above, the experiment contains many conditions. As a consequence, the data is quite big. Not all of this data has an interest to us for decoding, so we will keep only fMRI signals corresponding to faces or cats. We create a mask of the samples belonging to the condition; this mask is then applied to the fMRI data to restrict the classification to the face vs cat discrimination.
The input data will become much smaller (i.e. fMRI signal is shorter):
condition_mask = conditions.isin(["face", "cat"])
Because the data is in one single large 4D image, we need to use index_img to do the split easily.
from nilearn.image import index_img
fmri_niimgs = index_img(fmri_filename, condition_mask)
We apply the same mask to the targets
conditions = conditions[condition_mask]
conditions = conditions.to_numpy()
print(f"{conditions.shape=}")
conditions.shape=(216,)
Decoding with Support Vector Machine¶
As a decoder, we use a Support Vector Classifier with a linear kernel. We
first create it using by using Decoder.
from nilearn.decoding import Decoder
decoder = Decoder(
estimator="svc",
mask=mask_filename,
screening_percentile=100,
verbose=1,
)
Note
When viewing an Nilearn estimator in a notebook (or more generally on an HTML page like here) you get an expandable ‘Parameters’ section where the parameters that have different values from their default are highlighted in orange. If you are using a version of scikit-learn >= 1.8.0 you will also get access to the ‘docstring’ description of each parameter.
The decoder object is an object that can be fit (or trained) on data with labels, and then predict labels on data without.
We first fit it on the data.
Note
After fitting, the HTML representation of the estimator looks different than before before fitting.
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:172: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
We can then predict the labels from the data
prediction = decoder.predict(fmri_niimgs)
print(f"{prediction=}")
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
prediction=array(['face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'cat', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'face', 'face', 'face', 'face', 'face',
'face', 'face', 'face', 'cat', 'cat', 'cat', 'cat', 'cat', 'cat',
'cat', 'cat', 'cat'], dtype='<U4')
Note that for this classification task both classes contain the same number of samples (the problem is balanced). Then, we can use accuracy to measure the performance of the decoder. This is done by defining accuracy as the scoring. Let’s measure the prediction accuracy:
print((prediction == conditions).sum() / float(len(conditions)))
1.0
This prediction accuracy score is meaningless. Why?
Measuring prediction scores using cross-validation¶
The proper way to measure error rates or prediction accuracy is via cross-validation: leaving out some data and testing on it.
Manually leaving out data¶
Let’s leave out the 30 last data points during training, and test the prediction on these 30 last points:
fmri_niimgs_train = index_img(fmri_niimgs, slice(0, -30))
fmri_niimgs_test = index_img(fmri_niimgs, slice(-30, None))
conditions_train = conditions[:-30]
conditions_test = conditions[-30:]
decoder = Decoder(
estimator="svc",
mask=mask_filename,
screening_percentile=100,
verbose=1,
)
decoder.fit(fmri_niimgs_train, conditions_train)
prediction = decoder.predict(fmri_niimgs_test)
# The prediction accuracy is calculated on the test data: this is the accuracy
# of our model on examples it hasn't seen to examine how well the model perform
# in general.
predicton_accuracy = (prediction == conditions_test).sum() / float(
len(conditions_test)
)
print(f"Prediction Accuracy: {predicton_accuracy:.3f}")
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a04bdf90>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:213: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a04bdf90>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a04bf8e0>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Prediction Accuracy: 0.767
Implementing a KFold loop¶
We can manually split the data in train and test set repetitively in a KFold strategy by importing scikit-learn’s object:
from sklearn.model_selection import KFold
cv = KFold(n_splits=5)
for fold, (train, test) in enumerate(cv.split(conditions), start=1):
decoder = Decoder(
estimator="svc",
mask=mask_filename,
screening_percentile=100,
verbose=1,
)
decoder.fit(index_img(fmri_niimgs, train), conditions[train])
prediction = decoder.predict(index_img(fmri_niimgs, test))
predicton_accuracy = (prediction == conditions[test]).sum() / float(
len(conditions[test])
)
print(
f"CV Fold {fold:01d} | Prediction Accuracy: {predicton_accuracy:.3f}"
)
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a07b3370>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:243: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a07b3370>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a07b3c70>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
CV Fold 1 | Prediction Accuracy: 0.886
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a0c27af0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:243: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a0c27af0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a07b3370>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
CV Fold 2 | Prediction Accuracy: 0.767
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a057bfd0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:243: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a057bfd0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a057bca0>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
CV Fold 3 | Prediction Accuracy: 0.767
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a0728490>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:243: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a0728490>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a077b5e0>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
CV Fold 4 | Prediction Accuracy: 0.698
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a09c4fa0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:243: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a09c4fa0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a06f5720>
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
CV Fold 5 | Prediction Accuracy: 0.744
Cross-validation with the decoder¶
The decoder also implements a cross-validation loop by default and returns an array of shape (cross-validation parameters, n_folds). We can use accuracy score to measure its performance by defining accuracy as the scoring parameter.
n_folds = 5
decoder = Decoder(
estimator="svc",
mask=mask_filename,
cv=n_folds,
scoring="accuracy",
screening_percentile=100,
verbose=1,
)
decoder.fit(fmri_niimgs, conditions)
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:269: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.2s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Cross-validation pipeline can also be implemented manually. More details can be found on scikit-learn website.
Then we can check the best performing parameters per fold.
print(decoder.cv_params_["face"])
{'C': [np.float64(100.0), np.float64(100.0), np.float64(100.0), np.float64(100.0), np.float64(100.0)]}
Note
We can speed things up to use all the CPUs of our computer with the n_jobs parameter.
The best way to do cross-validation is to respect the structure of the experiment, for instance by leaving out full runs of acquisition.
The number of the run is stored in the CSV file giving the behavioral data. We have to apply our run mask, to select only cats and faces.
run_label = behavioral["chunks"][condition_mask]
The fMRI data is acquired by runs, and the noise is autocorrelated in a given run. Hence, it is better to predict across runs when doing cross-validation. To leave a run out, pass the cross-validator object to the cv parameter of decoder.
from sklearn.model_selection import LeaveOneGroupOut
cv = LeaveOneGroupOut()
decoder = Decoder(
estimator="svc",
mask=mask_filename,
cv=cv,
screening_percentile=100,
verbose=1,
)
decoder.fit(fmri_niimgs, conditions, groups=run_label)
print(f"{decoder.cv_scores_=}")
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:310: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32a03070a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
decoder.cv_scores_={np.str_('cat'): [1.0, 1.0, 1.0, 1.0, 0.9629629629629629, 0.8518518518518519, 0.9753086419753086, 0.40740740740740744, 0.9876543209876543, 1.0, 0.9259259259259259, 0.8765432098765432], np.str_('face'): [1.0, 1.0, 1.0, 1.0, 0.9629629629629629, 0.8518518518518519, 0.9753086419753086, 0.40740740740740744, 0.9876543209876543, 1.0, 0.9259259259259259, 0.8765432098765432]}
Inspecting the model weights¶
Finally, it may be useful to inspect and display the model weights.
Turning the weights into a nifti image¶
We retrieve the SVC discriminating weights
coef_ = decoder.coef_
print(f"{coef_=}")
coef_=array([[-3.89376986e-02, -1.87167729e-02, -3.23027195e-02,
-2.88745887e-02, 4.18696184e-02, 1.10743952e-02,
1.69997741e-02, -5.50955088e-02, -1.94205208e-02,
-3.51225099e-02, 1.08511262e-02, -1.28797423e-02,
-1.54678245e-02, -3.78907406e-02, -3.69170618e-02,
2.28086565e-02, 6.56421151e-03, -7.65762342e-03,
1.67105596e-02, -8.02140834e-03, 5.29515037e-02,
-8.17595412e-02, -6.36992710e-02, 2.41325974e-02,
4.59876650e-02, -2.22603255e-02, -1.77309904e-02,
2.22196614e-02, -9.53205485e-03, 5.76045109e-02,
2.14299326e-02, -9.14226015e-02, 4.03672822e-03,
-2.89275281e-02, -3.89030344e-02, -3.35114936e-02,
2.21398344e-03, 8.73136720e-03, -3.37415940e-02,
-2.41275113e-02, -6.81648777e-02, 1.65406057e-02,
2.70785487e-02, -6.56846220e-03, -1.21662117e-02,
5.47674579e-02, 8.13274509e-03, 3.60955536e-02,
-1.52762406e-02, 7.02912973e-02, 1.28100558e-03,
2.08009048e-02, -4.09952059e-03, 3.72429977e-02,
-3.77395048e-02, -1.03858150e-02, -2.38236832e-02,
-5.48880286e-02, 4.43028097e-02, -1.47419241e-01,
-2.34043169e-02, 1.87115139e-02, 6.65860369e-02,
-9.07603371e-02, -1.22035325e-02, -2.95641350e-03,
3.22091774e-02, -3.04053562e-02, 6.15345703e-02,
1.12248833e-02, 1.93776235e-02, -1.30542677e-02,
4.42975923e-02, -2.23065594e-02, 6.88146504e-02,
1.69390097e-02, 1.78947925e-02, 1.00277220e-02,
2.99186535e-02, -2.52170796e-02, 1.06156084e-02,
-6.31946743e-03, 2.21508966e-03, -2.23348652e-02,
1.42562010e-02, -1.53124062e-02, -1.98227330e-02,
-4.32638834e-02, -4.55124589e-02, 3.41589729e-02,
-2.79200305e-02, -2.80911524e-02, -3.70158715e-02,
-5.71451506e-02, -6.98951445e-02, 3.20168434e-03,
-8.35446208e-03, -3.37627439e-02, 3.04260940e-02,
8.68459167e-03, 6.19383083e-03, 5.94177586e-02,
9.07298257e-03, -1.48931135e-02, 1.43559839e-02,
-1.09027174e-02, 2.67698692e-02, 4.73787855e-02,
-2.96432475e-02, 3.09422495e-02, 1.57926782e-02,
-3.16720075e-02, -4.00106193e-02, -5.40260828e-02,
2.82611997e-02, -1.12101093e-02, -5.45402964e-02,
6.32177355e-02, -1.49996959e-02, 2.47544887e-03,
-4.56644346e-02, -1.83881582e-02, 1.19958557e-02,
-3.72172795e-02, -2.25521082e-03, 4.58656354e-02,
4.79164567e-02, 2.51828286e-03, -4.31721654e-02,
-5.35333960e-03, 5.76994993e-02, 7.40831874e-03,
-3.20590193e-02, 4.35711563e-03, 1.68303295e-02,
-2.92570229e-02, -2.24486338e-03, -8.30215673e-03,
-1.00012836e-02, 2.17135517e-02, -1.92617796e-03,
-1.33221660e-02, -2.80299734e-02, -1.75292853e-02,
-9.17800604e-03, -7.09935891e-03, -1.43032453e-02,
5.06832745e-02, -1.84812818e-02, -4.71508911e-02,
1.72568829e-02, -4.76641769e-02, -9.08861534e-04,
4.00770666e-02, 7.53995472e-02, 7.25613234e-03,
4.82604760e-02, 4.50556316e-02, 3.61202429e-02,
-8.16488031e-03, 1.95406536e-02, 3.57883767e-02,
4.89307594e-02, 3.82972595e-02, 6.23919165e-02,
6.13673638e-02, -1.68750750e-02, 1.66514155e-02,
3.35522672e-02, -1.80212840e-02, 4.46410723e-02,
-3.53244837e-02, -3.67291705e-02, -4.62256356e-03,
4.86829850e-02, 3.39667691e-02, 6.21707923e-03,
1.73612772e-02, 2.01698263e-02, 2.17097298e-02,
2.91414120e-02, 2.37777505e-02, 4.84697957e-02,
-9.22618522e-03, -2.82638002e-02, -2.13780793e-02,
1.80789649e-03, 4.79688079e-02, -9.78897373e-03,
1.11431089e-02, -1.65020719e-02, -2.89088464e-02,
2.42850285e-02, -1.22346375e-02, -2.92869940e-02,
-2.89847352e-02, -3.39532695e-02, -3.65284060e-03,
2.65323489e-02, 4.58039846e-02, -5.93382061e-02,
-2.13631521e-02, -3.09404447e-02, 5.50179410e-02,
-3.38816432e-02, 6.12624588e-03, 1.41483338e-02,
1.10215909e-02, 5.33811185e-02, -2.12339500e-02,
6.37418143e-03, -1.13075360e-02, -2.64226551e-02,
-2.22399606e-02, -5.31920281e-02, -3.98652571e-02,
-1.29727735e-01, -3.28091981e-02, -2.89709540e-02,
-9.13464583e-03, -7.28727129e-03, -3.71052354e-02,
-6.34906635e-02, 2.04392984e-03, -8.26795708e-02,
-6.71215666e-02, -2.29128101e-03, -2.33451802e-02,
1.77913311e-02, -8.74666902e-02, -2.76490071e-03,
-4.38275410e-02, -1.28051796e-02, 2.78033174e-02,
-4.32696561e-02, -3.22689030e-02, -2.28028685e-02,
-2.57413933e-02, 2.03623479e-02, -9.90248195e-03,
-3.15034446e-02, -1.81420293e-02, -1.12312128e-03,
-4.17432777e-02, -6.23480274e-02, 2.54773488e-04,
-6.73679604e-02, 6.53965842e-02, 1.06522047e-02,
2.21983654e-02, -1.98727956e-02, -1.85519244e-02,
4.05703009e-02, -3.02837349e-02, -8.10051881e-02,
-7.42459019e-02, -4.93850205e-02, -1.01770673e-02,
1.09407411e-02, -4.49253899e-02, 2.92748909e-02,
7.05307385e-03, 5.07540024e-03, -4.84046077e-03,
2.48727531e-03, 3.00655118e-02, -2.63085123e-03,
4.64686191e-03, 7.90209220e-02, 1.04858503e-02,
1.68080215e-02, -4.36719272e-02, -1.08855853e-02,
2.10241611e-02, -4.41964777e-02, 3.16495074e-03,
6.98672627e-02, 8.61631459e-02, 4.96234476e-02,
6.03902617e-03, 5.56494276e-02, -2.98921004e-02,
4.13036840e-03, -3.21952993e-02, -3.14991281e-02,
-5.31276728e-02, 2.67256957e-02, 3.14428204e-02,
6.67122363e-03, -1.28703337e-02, 2.20150792e-02,
5.68525274e-02, 2.25603340e-02, -2.04616944e-02,
5.10343199e-03, 2.85356318e-02, -1.81665046e-02,
-8.48436859e-03, -3.18823869e-02, -1.18499673e-02,
-4.10845665e-02, 3.11777908e-02, 9.63468178e-03,
-8.25916590e-03, -3.12229872e-02, 8.57642607e-03,
-9.70200226e-03, 1.32374604e-02, 4.06447582e-02,
8.23403014e-03, -3.27356893e-02, -4.33881099e-03,
-1.75530479e-02, 6.88842867e-03, 3.45132377e-02,
7.03298374e-02, 2.16785462e-02, 5.32228419e-03,
8.17564111e-02, 6.40062294e-02, -2.31136013e-03,
-1.17557036e-02, 1.75889230e-01, 3.18129161e-02,
-3.15887484e-02, 3.34027975e-02, 2.22783207e-02,
1.00231240e-02, -4.74914230e-02, -2.12757922e-02,
-3.98717625e-02, -6.04068676e-02, -4.65059292e-02,
1.03003229e-02, -3.05678631e-04, 1.80743109e-02,
-1.75453093e-02, -8.72591682e-02, 1.00662623e-01,
4.46117244e-03, 7.46870177e-02, -6.13411363e-02,
2.81704585e-02, -1.40977895e-02, 3.14637950e-02,
-1.63834626e-02, 3.66533517e-02, -5.15662400e-03,
1.45093561e-02, 6.35866683e-02, 2.34598869e-02,
8.81060718e-02, 6.15344663e-02, -1.39360761e-02,
2.07246667e-02, -3.15461110e-03, 5.15425280e-02,
-2.88767991e-02, 1.60263644e-02, 2.09702677e-02,
-3.29172614e-02, -2.59462054e-02, -5.60400830e-02,
-3.64627599e-02, 1.12881897e-02, 2.17266345e-02,
-1.51637346e-02, -7.82886072e-03, 2.42549982e-02,
9.47012775e-02, -2.63034890e-02, 1.17311759e-04,
-5.24176303e-03, 4.17988578e-02, 8.85680480e-02,
6.23640244e-03, 1.86599483e-02, 1.54629844e-02,
3.50541374e-03, 6.20605502e-03, -1.19790127e-02,
1.59526349e-02, 7.12125895e-03, -8.93193048e-02,
-3.54340932e-03, 1.23478274e-02, 3.03926654e-02,
-2.37295044e-02, -3.82792229e-02, -4.98746099e-02,
4.66894619e-02, -1.23292084e-02, -1.10332666e-02,
2.18106099e-02, 2.18719901e-02, 2.63537825e-02,
1.05280053e-02, 1.84617818e-02, 8.35939123e-04,
-6.65218031e-03, 3.49396792e-02, 1.49353763e-02,
-1.11598882e-02, 6.69099153e-03, -2.00058862e-02,
-3.99014976e-02, 3.01872551e-02, -1.09867485e-02,
-4.11780077e-02, 2.72051586e-02, 1.16427075e-02,
-1.55502682e-02, 3.27699490e-02, 3.95493200e-02,
8.48723373e-03, 2.19935342e-02, -9.88663757e-03,
-3.61421356e-02, -4.77020237e-02, 1.90072697e-02,
-5.58286645e-02, -3.31739505e-02, -2.24913565e-02,
-3.36175904e-02, -4.07355738e-02, 1.08861741e-02,
1.12813030e-02, 7.63145030e-02, 4.04803943e-03,
3.07014043e-02, 2.89177174e-02, 4.71614097e-03,
5.13383404e-02, -4.10364414e-02, 1.23249575e-03,
-2.50403510e-02, 5.85903989e-02, -1.04965675e-01,
-4.41704668e-02, 1.18518870e-02, -5.83202878e-02,
-4.82244144e-02, 9.17670145e-03, 1.03259671e-02,
-5.09184787e-03, -3.23390692e-02, -3.19385004e-02,
-1.53770284e-02, -5.21212793e-02, 1.55619685e-02,
2.93484319e-02, -1.92528526e-02, 1.76694108e-02,
2.67991563e-02, 5.76553591e-02, -1.38163391e-02,
2.60399634e-02, 1.50400953e-02, 1.27425411e-02,
-2.29243437e-02, -1.06664544e-02, 9.81940931e-03,
-4.77511782e-02, 1.64242840e-02]])
It’s a numpy array with only one coefficient per voxel:
print(f"{coef_.shape=}")
coef_.shape=(1, 464)
To get the Nifti image of these coefficients, we only need retrieve the coef_img_ in the decoder and select the class
coef_img = decoder.coef_img_["face"]
coef_img is now a NiftiImage. We can save the coefficients as a nii.gz file:
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_decoding_tutorial"
output_dir.mkdir(exist_ok=True, parents=True)
print(f"Output will be saved to: {output_dir}")
decoder.coef_img_["face"].to_filename(output_dir / "haxby_svc_weights.nii.gz")
Output will be saved to: /home/runner/work/nilearn/nilearn/examples/00_tutorials/results/plot_decoding_tutorial
Plotting the SVM weights¶
We can plot the weights, using the subject’s anatomical as a background
from nilearn.plotting import view_img
view_img(
decoder.coef_img_["face"],
bg_img=haxby_dataset.anat[0],
title="SVM weights",
dim=-1,
)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_decoding_tutorial.py:353: UserWarning:
Casting data from int16 to float32