Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.glm.first_level.FirstLevelModel¶
- class nilearn.glm.first_level.FirstLevelModel(t_r=None, slice_time_ref=0.0, hrf_model='glover', drift_model='cosine', high_pass=0.01, drift_order=1, fir_delays=None, min_onset=-24, mask_img=None, target_affine=None, target_shape=None, smoothing_fwhm=None, memory=None, memory_level=1, standardize=False, signal_scaling=0, noise_model='ar1', verbose=0, n_jobs=1, minimize_memory=True, subject_label=None, random_state=None)[source]¶
Implement the General Linear Model for single run fMRI data.
- Parameters:
- t_r
floator None, default=None This parameter indicates repetition times of the experimental runs. In seconds. It is necessary to correctly consider times in the design matrix. This parameter is also passed to
nilearn.signal.clean. Please see the related documentation for details.Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- slice_time_ref
float, default=0.0 This parameter indicates the time of the reference slice used in the slice timing preprocessing step of the experimental runs. It is expressed as a fraction of the
t_r(repetition time), so it can have values between 0. and 1.Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- hrf_model
str, function,listof functions, or None This parameter defines the HRF model to be used. It can be a string if you are passing the name of a model implemented in Nilearn. Valid names are:
"spm + derivative": SPM model plus its time derivative. This gives 2 regressors. Seespm_hrf, andspm_time_derivative."spm + derivative + dispersion": Same as above plus dispersion derivative. This gives 3 regressors. Seespm_hrf,spm_time_derivative, andspm_dispersion_derivative."glover": This corresponds to the Glover HRF. Seeglover_hrf."glover + derivative": The Glover HRF + time derivative. This gives 2 regressors. Seeglover_hrf, andglover_time_derivative."glover"+ derivative + dispersion": Same as above plus dispersion derivative. This gives 3 regressors. Seeglover_hrf,glover_time_derivative, andglover_dispersion_derivative."fir": Finite impulse response basis. This is a set of delayed dirac models.
It can also be a custom model. In this case, a function should be provided for each regressor. Each function should behave as the other models implemented within Nilearn. That is, it should take both
t_randoversamplingas inputs and return a sample numpy array of appropriate shape.Note
It is expected that
"spm"standard and"glover"models would not yield large differences in most cases.Note
In case of
"glover"and"spm"models, the derived regressors are orthogonalized with respect to the main one.Default=’glover’.
Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- drift_model
str, default=’cosine’ This parameter specifies the desired drift model for the design matrices. It can be ‘polynomial’, ‘cosine’ or None.
Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- high_pass
float, default=0.01 This parameter specifies the cut frequency of the high-pass filter in Hz for the design matrices. Used only if drift_model is ‘cosine’.
Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- drift_order
int, default=1 This parameter specifies the order of the drift model (in case it is polynomial) for the design matrices.
Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- fir_delaysarray of shape(n_onsets),
listor None, default=None Will be set to
[0]ifNoneis passed. In case of FIR design, yields the array of delays used in the FIR model, in scans.Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- min_onset
float, default=-24 This parameter specifies the minimal onset relative to the design (in seconds). Events that start before (slice_time_ref * t_r + min_onset) are not considered.
Warning
This parameter is ignored by fit() if design matrices are passed at fit time.
- mask_imgNiimg-like, NiftiMasker,
SurfaceImage,SurfaceMasker, False or None, default=None Mask to be used on data. If an instance of masker is passed, then its mask will be used. If None is passed, the mask will be computed automatically by a NiftiMasker or
SurfaceMaskerwith default parameters. If False is given then the data will not be masked. In the case of surface analysis, passing None or False will lead to no masking.- target_affine3x3 or a 4x4 array-like, or None, default=None
If specified, the image is resampled corresponding to this new affine.
Note
This parameter is passed to
nilearn.image.resample_img.- target_shape
tupleorlistor None, default=None If specified, the image will be resized to match this new shape. len(target_shape) must be equal to 3.
Note
If target_shape is specified, a target_affine of shape (4, 4) must also be given.
Note
This parameter is passed to
nilearn.image.resample_img.- smoothing_fwhm
floatorintor None, optional. If smoothing_fwhm is not None, it gives the full-width at half maximum in millimeters of the spatial smoothing to apply to the signal.
- memoryNone, instance of
joblib.Memory,str, orpathlib.Path, default=None Used to cache the masking process. By default, no caching is done. If a
stris given, it is the path to the caching directory.- memory_level
int, default=0 Rough estimator of the amount of memory used by caching. Higher value means more memory for caching. Zero means no caching.
- standardizeany of: ‘zscore_sample’, ‘zscore’, ‘psc’, True, False or None; default=False
Strategy to standardize the signal:
'zscore_sample': The signal is z-scored. Timeseries are shifted to zero mean and scaled to unit variance. Uses sample std.'psc': Timeseries are shifted to zero mean value and scaled to percent signal change (as compared to original mean signal).True: The signal is z-scored (same as option zscore). Timeseries are shifted to zero mean and scaled to unit variance.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Truewill be replaced by'zscore_sample'.False: Do not standardize the data.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Falsewill be replaced byNone.
Deprecated since Nilearn 0.13.0: The default will be changed to
Nonein version 0.15.0.- signal_scalingFalse,
intor (int, int), default=0 If not False, fMRI signals are scaled to the mean value of scaling_axis given, which can be 0, 1 or (0, 1). 0 refers to mean scaling each voxel with respect to time, 1 refers to mean scaling each time point with respect to all voxels & (0, 1) refers to scaling with respect to voxels and time, which is known as grand mean scaling. Incompatible with standardize (
standardize=Noneis enforced whensignal_scalingis not False).- noise_model{‘ar1’, ‘ols’}, default=’ar1’
The temporal variance model.
- verbose
boolorint, default=0 Verbosity level (
0orFalsemeans no message). If 0, prints nothing If 1, prints progress by computation of each run. If 2, prints timing details of masker and GLM. If 3, prints masker computation details.- n_jobs
int, default=1 The number of CPUs to use to do the computation. -1 means ‘all CPUs’.
- minimize_memory
bool, default=True Gets rid of some variables on the model fit results that are not necessary for contrast computation and would only be useful for further inspection of model details. This has an important impact on memory consumption.
- subject_label
stror None, default=None This id will be used to identify a FirstLevelModel when passed to a SecondLevelModel object.
- random_state
intor numpy.random.RandomState, default=None. Random state seed to sklearn.cluster.KMeans for autoregressive models of order at least 2 (‘ar(N)’ with n >= 2).
Added in Nilearn 0.9.1.
- t_r
- Attributes:
- design_matrices_
listofpandas.DataFrame Design matrices used to fit the GLM.
- fir_delays_array of shape(n_onsets),
list - labels_array of shape
(n_elements_,) a map of values on voxels / vertices used to identify the corresponding model
- masker_
NiftiMaskerorSurfaceMasker Masker used to filter and mask data during fit. If
NiftiMaskerorSurfaceMaskeris given inmask_imgparameter, this is a copy of it. Otherwise, a masker is created using the value ofmask_imgand other NiftiMasker/SurfaceMasker related parameters as initialization.- memory_joblib memory cache
- n_elements_
int The number of voxels or vertices in the mask.
Added in Nilearn 0.12.1.
- results_
dict, with keys corresponding to the different labels values. Values are SimpleRegressionResults corresponding to the voxels, if minimize_memory is True, RegressionResults if minimize_memory is False
- standardize_any of: ‘zscore_sample’, ‘zscore’, ‘psc’, or None
This value may differ from the
standardizeparameters as it is set toNonewhensignal_scalingis not False.
- design_matrices_
- __init__(t_r=None, slice_time_ref=0.0, hrf_model='glover', drift_model='cosine', high_pass=0.01, drift_order=1, fir_delays=None, min_onset=-24, mask_img=None, target_affine=None, target_shape=None, smoothing_fwhm=None, memory=None, memory_level=1, standardize=False, signal_scaling=0, noise_model='ar1', verbose=0, n_jobs=1, minimize_memory=True, subject_label=None, random_state=None)[source]¶
- compute_contrast(contrast_def, stat_type=None, output_type='z_score')[source]¶
Generate different outputs corresponding to the contrasts provided e.g. z_map, t_map, effects and variance.
In multi-run case, outputs the fixed effects map.
- Parameters:
- contrast_def
stror array of shape (n_col) orlistof (stror array of shape (n_col)) where
n_colis the number of columns of the design matrix, (one array per run). If only one array is provided when there are several runs, it will be assumed that the same contrast is desired for all runs. One can use the name of the conditions as they appear in the design matrix of the fitted model combined with operators +- and combined with numbers with operators +-*/. In this case, the string defining the contrasts must be a valid expression for compatibility withpandas.DataFrame.eval.- stat_type{‘t’, ‘F’}, default=None
Type of the contrast.
- output_type
str, default=’z_score’ Type of the output map. Can be ‘z_score’, ‘stat’, ‘p_value’, ‘effect_size’, ‘effect_variance’ or ‘all’.
- contrast_def
- Returns:
- outputNifti1Image,
SurfaceImage, ordict The desired output image(s). If
output_type == 'all', then the output is a dictionary of images, keyed by the type of image.
- outputNifti1Image,
- fit(run_imgs, events=None, confounds=None, sample_masks=None, design_matrices=None, bins=100)[source]¶
Fit the GLM.
For each run: 1. create design matrix X 2. do a masker job: fMRI_data -> Y 3. fit regression to (Y, X)
Warning
If design_matrices are passed to fit(), then the following attributes are ignored:
drift_model,drift_order,fir_delays,high_pass,hrf_model,min_onset,slice_time_ref,t_r.- Parameters:
- run_imgsNiimg-like object,
listortupleof Niimg-like objects, SurfaceImage object, orlistortupleofSurfaceImage Data on which the GLM will be fitted. If this is a list, the affine is considered the same for all.
Warning
If the FirstLevelModel object was instantiated with a
mask_img, thenrun_imgsmust be compatible withmask_img. For example, ifmask_imgis anilearn.maskers.NiftiMaskerinstance or a Niimng-like object, thenrun_imgsmust be a Niimg-like object, alistor atupleof Niimg-like objects. Ifmask_imgis aSurfaceMaskerorSurfaceImageinstance, thenrun_imgsmust be aSurfaceImage, alistor atupleofSurfaceImage.- events
pandas.DataFrameorpandas.Seriesorstrorpathlib.Pathto a TSV file, orlistofpandas.DataFrame,strorpathlib.Pathto a TSV file, or None, default=None fMRI events used to build design matrices. One events object expected per run_img. Ignored in case designs is not None. If string, then a path to a csv or tsv file is expected. See
make_first_level_design_matrixfor details on the required content of events files.Warning
This parameter is ignored if design_matrices are passed.
- confounds
pandas.DataFrame,numpy.ndarrayorstrorlistofpandas.DataFrame,numpy.ndarrayorstr, default=None Each column in a DataFrame corresponds to a confound variable to be included in the regression model of the respective run_img. The number of rows must match the number of volumes in the respective run_img. Ignored in case designs is not None. If string, then a path to a csv file is expected.
Warning
This parameter is ignored if design_matrices are passed.
- sample_masksarray_like, or
listof array_like, default=None shape of array: (number of scans - number of volumes remove) Indices of retained volumes. Masks the niimgs along time/fourth dimension to perform scrubbing (remove volumes with high motion) and/or remove non-steady-state volumes.
Added in Nilearn 0.9.2.
- design_matrices
pandas.DataFrameorstrorpathlib.Pathto a CSV or TSV file, orlistofpandas.DataFrame,strorpathlib.Pathto a CSV or TSV file, or None, default=None Design matrices that will be used to fit the GLM. If given it takes precedence over events and confounds.
- bins
int, default=100 Maximum number of discrete bins for the AR coef histogram. If an autoregressive model with order greater than one is specified then adaptive quantification is performed and the coefficients will be clustered via K-means with bins number of clusters.
- run_imgsNiimg-like object,
- generate_report(contrasts=None, first_level_contrast=None, title=None, bg_img='MNI152TEMPLATE', threshold=None, alpha=0.001, cluster_threshold=0, height_control='fpr', two_sided=False, min_distance=8.0, plot_type='slice', cut_coords=None, display_mode=None, report_dims=(1600, 800))[source]¶
Return a
HTMLReportwhich shows all important aspects of a fitted GLM.The
HTMLReportcan be opened in a browser, displayed in a notebook, or saved to disk as a standalone HTML file.The GLM must be fitted and have the computed design matrix(ces).
- Parameters:
- contrasts
dictwithstr- ndarray key-value pairs orstrorlistofstror ndarray orlistof ndarray, Default=None Contrasts information for a first or second level model.
Example:
Contrasts are passed to
contrast_deffor FirstLevelModel (nilearn.glm.first_level.FirstLevelModel.compute_contrast) & second_level_contrast for SecondLevelModel (nilearn.glm.second_level.SecondLevelModel.compute_contrast)- first_level_contrast
strornumpy.ndarrayof shape (n_col) with respect toFirstLevelModelor None, default=None When the model is a
SecondLevelModel:in case a
listofFirstLevelModelwas provided assecond_level_input, we have to provide a contrast to apply to the first level models to get the corresponding list of images desired, that would be tested at the second level,in case a
DataFramewas provided assecond_level_inputthis is the map name to extract from theDataFramemap_namecolumn. (it has to be a ‘t’ contrast).
When the model is a
FirstLevelModel: This parameter is ignored.Added in Nilearn 0.12.0.
- title
stror None, default=None If string, represents the web page’s title and primary heading, model type is sub-heading. If None, page titles and headings are autogenerated using contrast names.
- bg_imgNiimg-like object, default=’MNI152TEMPLATE’
See Input and output: neuroimaging data representation. The background image for mask and stat maps to be plotted on upon. To turn off background image, just pass “bg_img=None”.
- threshold
floatorintor None, default=None Cluster forming threshold in same scale as stat_img (either a t-scale or z-scale value). Used only if height_control is None. If
thresholdis set to None whenheight_controlis None,thresholdwill be set to 3.09.Note
When
two_sidedis True:'threshold'cannot be negative.The given value should be within the range of minimum and maximum intensity of the input image. All intensities in the interval
[-threshold, threshold]will be set to zero.When
two_sidedis False:If the threshold is negative:
It should be greater than the minimum intensity of the input data. All intensities greater than or equal to the specified threshold will be set to zero. All other intensities keep their original values.
If the threshold is positive:
It should be less than the maximum intensity of the input data. All intensities less than or equal to the specified threshold will be set to zero. All other intensities keep their original values.
- alpha
float, default=0.001 Number controlling the thresholding (either a p-value or q-value). Its actual meaning depends on the height_control parameter. This function translates alpha to a z-scale threshold.
- cluster_threshold
int, default=0 Cluster size threshold. Sets of connected voxels / vertices (clusters) with size smaller than this number will be removed.
- cluster_threshold
- height_control
stror None, default=’fpr’ false positive control meaning of cluster forming threshold: ‘fpr’ or ‘fdr’ or ‘bonferroni’ or None.
- two_sided
bool, default=False Whether to employ two-sided thresholding or to evaluate positive values only.
- min_distance
float, default=8.0 For display purposes only. Minimum distance between subpeaks in mm.
- plot_type
str, {‘slice’, ‘glass’}, default=’slice’ Specifies the type of plot to be drawn for the statistical maps.
- cut_coordsNone, allowed types depend on the
display_mode, optional The world coordinates of the point where the cut is performed.
If
display_modeis'ortho'or'tiled', this must be a 3-sequence offloatorint:(x, y, z).If
display_modeis'xz','yz'or'yx', this must be a 2-sequence offloatorint:(x, z),(y, z)or(x, y).If
display_modeis"x","y", or"z", this can be:If
display_modeis'mosaic', this can be:an
intin which case it specifies the number of cuts to perform in each direction"x","y","z".a 3-sequence of
floatorintin which case it specifies the number of cuts to perform in each direction"x","y","z"separately.dict<str: 1Dndarray> in which case keys are the directions (‘x’, ‘y’, ‘z’) and the values are sequences holding the cut coordinates.
If
Noneis given, the cuts are calculated automatically.
Note
cut_coordswill not be used whenplot_type='glass'.- display_mode
str, default=None Default is ‘z’ if plot_type is ‘slice’; ‘ortho’ if plot_type is ‘glass’.
Choose the direction of the cuts: ‘x’ - sagittal, ‘y’ - coronal, ‘z’ - axial, ‘l’ - sagittal left hemisphere only, ‘r’ - sagittal right hemisphere only, ‘ortho’ - three cuts are performed in orthogonal directions.
Possible values are: ‘ortho’, ‘x’, ‘y’, ‘z’, ‘xz’, ‘yx’, ‘yz’, ‘l’, ‘r’, ‘lr’, ‘lzr’, ‘lyr’, ‘lzry’, ‘lyrz’.
- report_dimsSequence[
int,int], default=(1600, 800) Specifies width, height (in pixels) of report window within a notebook. Only applicable when inserting the report into a Jupyter notebook. Can be set after report creation using report.width, report.height.
- contrasts
- Returns:
- report_text
HTMLReport Contains the HTML code for the GLM report.
- report_text
- get_metadata_routing()¶
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- property mask_img_¶
Return mask image using during fit.
- property predicted¶
Transform voxelwise predicted values to the same shape as the input Nifti1Image(s).
- Returns:
- outputlist
A list of Nifti1Image(s).
- property r_square¶
Transform voxelwise r-squared values to the same shape as the input Nifti1Image(s).
- Returns:
- outputlist
A list of Nifti1Image(s).
- property residuals¶
Transform voxelwise residuals to the same shape as the input Nifti1Image(s).
- Returns:
- outputlist
A list of Nifti1Image(s).
- set_fit_request(*, bins='$UNCHANGED$', confounds='$UNCHANGED$', design_matrices='$UNCHANGED$', events='$UNCHANGED$', run_imgs='$UNCHANGED$', sample_masks='$UNCHANGED$')¶
Configure whether metadata should be requested to be passed to the
fitmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- binsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
binsparameter infit.- confoundsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
confoundsparameter infit.- design_matricesstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
design_matricesparameter infit.- eventsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
eventsparameter infit.- run_imgsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
run_imgsparameter infit.- sample_masksstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
sample_masksparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using nilearn.glm.first_level.FirstLevelModel¶
Intro to GLM Analysis: a single-run, single-subject fMRI dataset
Decoding of a dataset after GLM fit for signal extraction
Analysis of an fMRI dataset with a Finite Impule Response (FIR) model
First level analysis of a complete BIDS dataset from openneuro
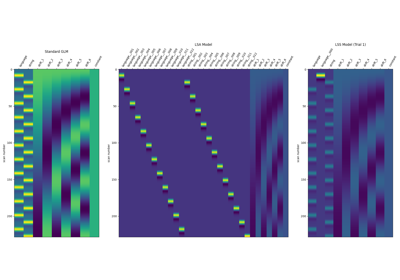
Beta-Series Modeling for Task-Based Functional Connectivity and Decoding
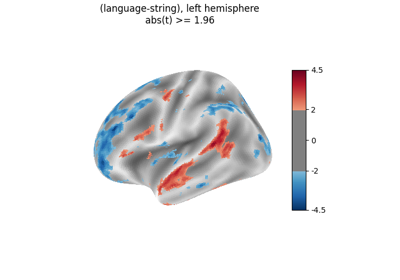
Surface-based dataset first and second level analysis of a dataset