Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Single-subject data (two runs) in native space¶
The example shows the analysis of an SPM dataset, with two conditions: viewing a face image or a scrambled face image.
This example takes a lot of time because the input are lists of 3D images sampled in different positions (encoded by different affine functions).
See also
For more information see the dataset description.
Fetch and inspect the data¶
Fetch the SPM multimodal_faces data.
from nilearn.datasets import fetch_spm_multimodal_fmri
subject_data = fetch_spm_multimodal_fmri()
[fetch_spm_multimodal_fmri] Dataset created in
/home/runner/nilearn_data/spm_multimodal_fmri
[fetch_spm_multimodal_fmri] Missing 390 functional scans for session 1.
[fetch_spm_multimodal_fmri] Data absent, downloading...
[fetch_spm_multimodal_fmri] Downloading data from
https://www.fil.ion.ucl.ac.uk/spm/download/data/mmfaces/multimodal_fmri.zip ...
[fetch_spm_multimodal_fmri] Downloaded 3358720 of 134263085 bytes (2.5%%,
40.2s remaining)
[fetch_spm_multimodal_fmri] Downloaded 8503296 of 134263085 bytes (6.3%%,
30.2s remaining)
[fetch_spm_multimodal_fmri] Downloaded 13279232 of 134263085 bytes (9.9%%,
27.7s remaining)
[fetch_spm_multimodal_fmri] Downloaded 18374656 of 134263085 bytes (13.7%%,
25.5s remaining)
[fetch_spm_multimodal_fmri] Downloaded 23658496 of 134263085 bytes (17.6%%,
23.6s remaining)
[fetch_spm_multimodal_fmri] Downloaded 28811264 of 134263085 bytes (21.5%%,
22.1s remaining)
[fetch_spm_multimodal_fmri] Downloaded 34127872 of 134263085 bytes (25.4%%,
20.7s remaining)
[fetch_spm_multimodal_fmri] Downloaded 39297024 of 134263085 bytes (29.3%%,
19.5s remaining)
[fetch_spm_multimodal_fmri] Downloaded 44711936 of 134263085 bytes (33.3%%,
18.1s remaining)
[fetch_spm_multimodal_fmri] Downloaded 49922048 of 134263085 bytes (37.2%%,
17.0s remaining)
[fetch_spm_multimodal_fmri] Downloaded 55623680 of 134263085 bytes (41.4%%,
15.7s remaining)
[fetch_spm_multimodal_fmri] Downloaded 61399040 of 134263085 bytes (45.7%%,
14.3s remaining)
[fetch_spm_multimodal_fmri] Downloaded 67518464 of 134263085 bytes (50.3%%,
12.9s remaining)
[fetch_spm_multimodal_fmri] Downloaded 74358784 of 134263085 bytes (55.4%%,
11.4s remaining)
[fetch_spm_multimodal_fmri] Downloaded 81616896 of 134263085 bytes (60.8%%,
9.7s remaining)
[fetch_spm_multimodal_fmri] Downloaded 89997312 of 134263085 bytes (67.0%%,
7.9s remaining)
[fetch_spm_multimodal_fmri] Downloaded 94699520 of 134263085 bytes (70.5%%,
7.2s remaining)
[fetch_spm_multimodal_fmri] Downloaded 98689024 of 134263085 bytes (73.5%%,
6.5s remaining)
[fetch_spm_multimodal_fmri] Downloaded 102400000 of 134263085 bytes (76.3%%,
6.0s remaining)
[fetch_spm_multimodal_fmri] Downloaded 106430464 of 134263085 bytes (79.3%%,
5.3s remaining)
[fetch_spm_multimodal_fmri] Downloaded 109993984 of 134263085 bytes (81.9%%,
4.7s remaining)
[fetch_spm_multimodal_fmri] Downloaded 113131520 of 134263085 bytes (84.3%%,
4.1s remaining)
[fetch_spm_multimodal_fmri] Downloaded 116400128 of 134263085 bytes (86.7%%,
3.5s remaining)
[fetch_spm_multimodal_fmri] Downloaded 119873536 of 134263085 bytes (89.3%%,
2.9s remaining)
[fetch_spm_multimodal_fmri] Downloaded 123355136 of 134263085 bytes (91.9%%,
2.2s remaining)
[fetch_spm_multimodal_fmri] Downloaded 126844928 of 134263085 bytes (94.5%%,
1.5s remaining)
[fetch_spm_multimodal_fmri] Downloaded 130342912 of 134263085 bytes (97.1%%,
0.8s remaining)
[fetch_spm_multimodal_fmri] Downloaded 133824512 of 134263085 bytes (99.7%%,
0.1s remaining)
[fetch_spm_multimodal_fmri] ...done. (29 seconds, 0 min)
[fetch_spm_multimodal_fmri] Extracting data from
/home/runner/nilearn_data/spm_multimodal_fmri/sub001/multimodal_fmri.zip...
[fetch_spm_multimodal_fmri] .. done.
[fetch_spm_multimodal_fmri] Downloading data from
https://www.fil.ion.ucl.ac.uk/spm/download/data/mmfaces/multimodal_smri.zip ...
[fetch_spm_multimodal_fmri] Downloaded 4349952 of 6852766 bytes (63.5%%, 0.6s
remaining)
[fetch_spm_multimodal_fmri] ...done. (2 seconds, 0 min)
[fetch_spm_multimodal_fmri] Extracting data from
/home/runner/nilearn_data/spm_multimodal_fmri/sub001/multimodal_smri.zip...
[fetch_spm_multimodal_fmri] .. done.
Let’s inspect one of the event files before using them.
import pandas as pd
events = [subject_data.events1, subject_data.events2]
events_dataframe = pd.read_csv(events[0], sep="\t")
events_dataframe["trial_type"].value_counts()
trial_type
scrambled 86
faces 64
Name: count, dtype: int64
We can confirm there are only 2 conditions in the dataset.
from nilearn.plotting import plot_event, show
plot_event(events)
show()
Resample the images:
this is achieved by the concat_imgs function of Nilearn.
import warnings
from nilearn.image import concat_imgs, mean_img, resample_img
# Avoid getting too many warnings due to resampling
with warnings.catch_warnings():
warnings.simplefilter("ignore")
fmri_img = [
concat_imgs(subject_data.func1, auto_resample=True),
concat_imgs(subject_data.func2, auto_resample=True),
]
affine, shape = fmri_img[0].affine, fmri_img[0].shape
print("Resampling the second image (this takes time)...")
fmri_img[1] = resample_img(fmri_img[1], affine, shape[:3])
Resampling the second image (this takes time)...
Let’s create mean image for display purposes.
Fit the model¶
Fit the GLM for the 2 runs by specifying a FirstLevelModel and then fitting it.
# Sample at the beginning of each acquisition.
slice_time_ref = 0.0
# We use a discrete cosine transform to model signal drifts.
drift_model = "cosine"
# The cutoff for the drift model is 0.01 Hz.
high_pass = 0.01
# The hemodynamic response function
hrf_model = "spm + derivative"
from nilearn.glm.first_level import FirstLevelModel
print("Fitting a GLM")
fmri_glm = FirstLevelModel(
smoothing_fwhm=None,
t_r=subject_data.t_r,
hrf_model=hrf_model,
drift_model=drift_model,
high_pass=high_pass,
verbose=1,
)
fmri_glm = fmri_glm.fit(fmri_img, events=events)
Fitting a GLM
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f5d7faefbe0>
[FirstLevelModel.fit] Computing mask
[FirstLevelModel.fit] Resampling mask
[FirstLevelModel.fit] Finished fit
[FirstLevelModel.fit] Computing run 1 out of 2 runs (go take a coffee, a big
one).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f5d7faefbe0>
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 0 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 1 seconds.
[FirstLevelModel.fit] Computing run 2 out of 2 runs (2 seconds remaining).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f5d98472620>
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 0 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 1 seconds.
[FirstLevelModel.fit] Computation of 2 runs done in 4 seconds.
View the results¶
Now we can compute contrast-related statistical maps (in z-scale), and plot them.
from nilearn.plotting import plot_stat_map
print("Computing contrasts")
Computing contrasts
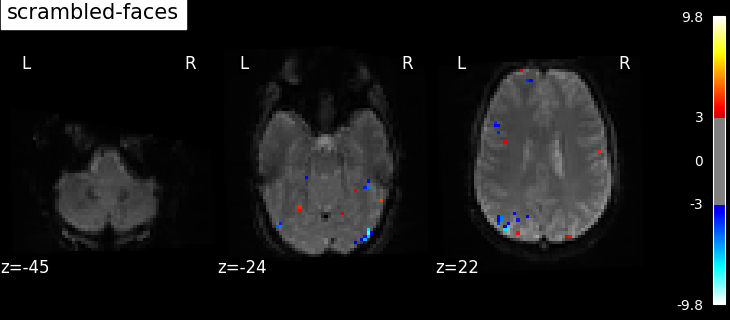
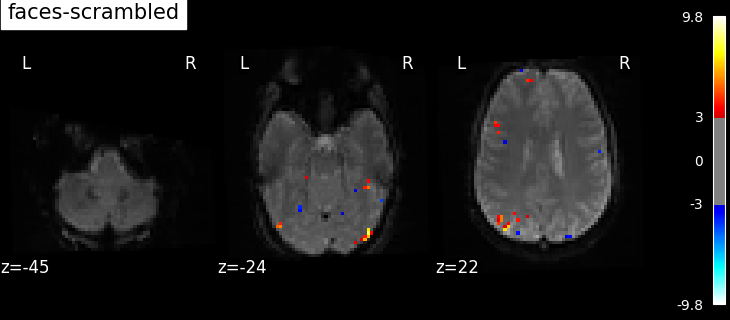
We actually want more interesting contrasts. The simplest contrast just makes the difference between the two main conditions. We define the two opposite versions to run one-tailed t-tests.
contrasts = ["faces - scrambled", "scrambled - faces"]
Let’s store common parameters for all plots.
We plot the contrasts values overlaid on the mean fMRI image and we will use the z-score values as transparency, with any voxel with | Z-score | > 3 being fully opaque and any voxel with 0 < | Z-score | < 1.96 being partly transparent.
plot_param = {
"vmin": 0,
"display_mode": "z",
"cut_coords": 3,
"black_bg": True,
"bg_img": mean_image,
"cmap": "inferno",
"transparency_range": [0, 3],
}
# Iterate on contrasts to compute and plot them.
for contrast_id in contrasts:
print(f"\tcontrast id: {contrast_id}")
results = fmri_glm.compute_contrast(contrast_id, output_type="all")
plot_stat_map(
results["stat"],
title=contrast_id,
transparency=results["z_score"],
**plot_param,
)
contrast id: faces - scrambled
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_spm_multimodal_faces.py:134: RuntimeWarning: The same contrast will be used for all 2 runs. If the design matrices are not the same for all runs, (for example with different column names or column order across runs) you should pass contrast as an expression using the name of the conditions as they appear in the design matrices.
results = fmri_glm.compute_contrast(contrast_id, output_type="all")
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
contrast id: scrambled - faces
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_spm_multimodal_faces.py:134: RuntimeWarning: The same contrast will be used for all 2 runs. If the design matrices are not the same for all runs, (for example with different column names or column order across runs) you should pass contrast as an expression using the name of the conditions as they appear in the design matrices.
results = fmri_glm.compute_contrast(contrast_id, output_type="all")
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
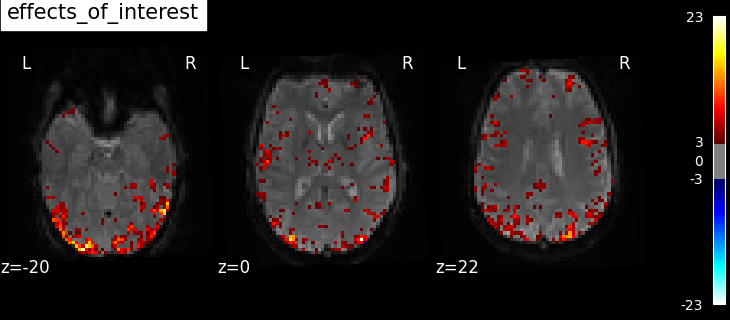
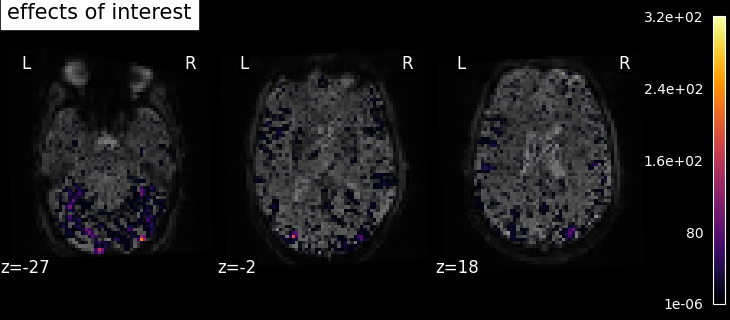
We also define the effects of interest contrast, a 2-dimensional contrasts spanning the two conditions.
import numpy as np
contrasts = np.eye(2)
results = fmri_glm.compute_contrast(contrasts, output_type="all")
plot_stat_map(
results["stat"],
title="effects of interest",
transparency=results["z_score"],
**plot_param,
)
show()
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_spm_multimodal_faces.py:151: RuntimeWarning: The same contrast will be used for all 2 runs. If the design matrices are not the same for all runs, (for example with different column names or column order across runs) you should pass contrast as an expression using the name of the conditions as they appear in the design matrices.
results = fmri_glm.compute_contrast(contrasts, output_type="all")
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_spm_multimodal_faces.py:151: UserWarning: F contrasts should have 20 columns, but it has only 2. The rest of the contrast was padded with zeros.
results = fmri_glm.compute_contrast(contrasts, output_type="all")
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_spm_multimodal_faces.py:151: UserWarning: Running approximate fixed effects on F statistics.
results = fmri_glm.compute_contrast(contrasts, output_type="all")
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
Based on the resulting maps we observe that the analysis results in wide activity for the ‘effects of interest’ contrast, showing the implications of large portions of the visual cortex in the conditions. By contrast, the differential effect between “faces” and “scrambled” involves sparser, more anterior and lateral regions. It also displays some responses in the frontal lobe.
Total running time of the script: (1 minutes 24.802 seconds)
Estimated memory usage: 920 MB