Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Computing a connectome with sparse inverse covariance¶
This example constructs a functional connectome using the sparse inverse covariance.
We use the MSDL atlas
of functional regions in movie watching, and the
NiftiMapsMasker to extract time series.
Note that the inverse covariance (or precision) contains values that can be linked to negated partial correlations, so we negated it for display.
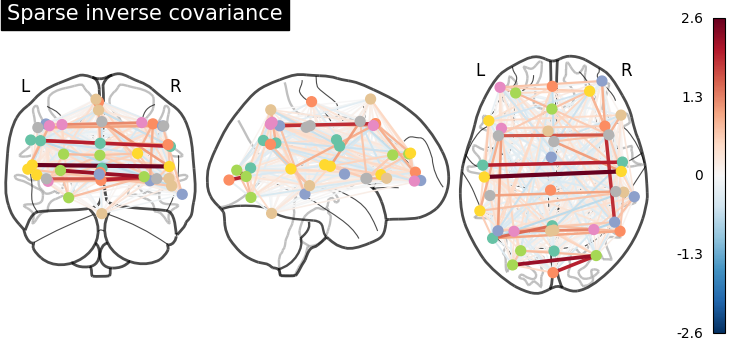
As the MSDL atlas comes with (x, y, z) MNI coordinates for the different regions, we can visualize the matrix as a graph of interaction in a brain. To avoid having too dense a graph, we represent only the 20% edges with the highest values.
Retrieve the atlas and the data¶
from nilearn.datasets import fetch_atlas_msdl, fetch_development_fmri
atlas = fetch_atlas_msdl()
# Loading atlas image stored in 'maps'
atlas_filename = atlas["maps"]
# Loading atlas data stored in 'labels'
labels = atlas["labels"]
# Loading the functional datasets
data = fetch_development_fmri(n_subjects=1)
# print basic information on the dataset
print(f"First subject functional nifti images (4D) are at: {data.func[0]}")
[fetch_atlas_msdl] Dataset found in /home/runner/nilearn_data/msdl_atlas
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
First subject functional nifti images (4D) are at: /home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Extract time series¶
from nilearn.maskers import NiftiMapsMasker
masker = NiftiMapsMasker(
maps_img=atlas_filename,
standardize_confounds=True,
memory="nilearn_cache",
memory_level=1,
verbose=1,
)
time_series = masker.fit_transform(data.func[0], confounds=data.confounds)
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_inverse_covariance_connectome.py:54: FutureWarning:
boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f6f321c5e70>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': False,
'dtype': None,
'high_pass': None,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': None,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': None,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_inverse_covariance_connectome.py:54: FutureWarning:
boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
_______________________________________________filter_and_extract - 0.8s, 0.0min
Compute the sparse inverse covariance¶
from sklearn.covariance import GraphicalLassoCV
estimator = GraphicalLassoCV(verbose=True)
estimator.fit(time_series)
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.2s finished
[GraphicalLassoCV] Done refinement 1 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.3s finished
[GraphicalLassoCV] Done refinement 2 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.5s finished
[GraphicalLassoCV] Done refinement 3 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.5s finished
[GraphicalLassoCV] Done refinement 4 out of 4: 1s
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/_methods.py:191: RuntimeWarning:
invalid value encountered in subtract
Display the connectome matrix¶
from nilearn.plotting import (
plot_connectome,
plot_matrix,
show,
view_connectome,
)
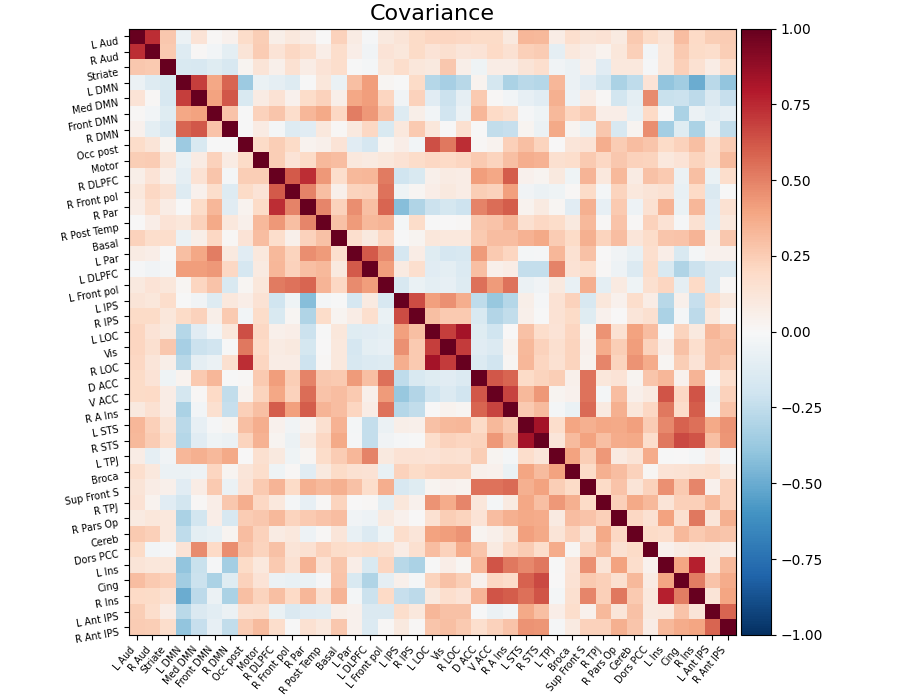
# Display the covariance
# The covariance can be found at estimator.covariance_
plot_matrix(
estimator.covariance_,
labels=labels,
figure=(9, 7),
vmax=1,
vmin=-1,
title="Covariance",
)
<matplotlib.image.AxesImage object at 0x7f6f421e3370>
And now display the corresponding graph¶
coords = atlas.region_coords
plot_connectome(estimator.covariance_, coords, title="Covariance")
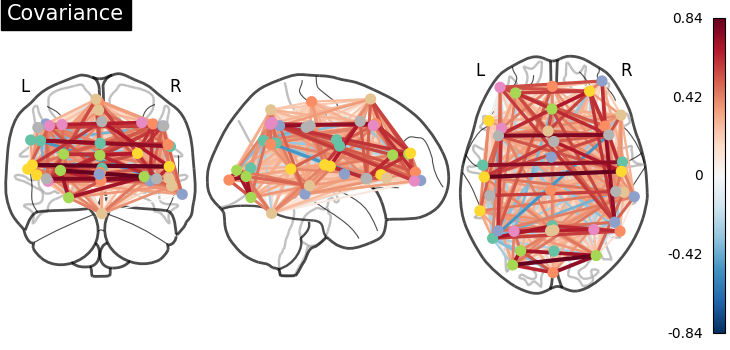
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f6f3f226c50>
Display the sparse inverse covariance¶
we negate it to get partial correlations
plot_matrix(
-estimator.precision_,
labels=labels,
figure=(9, 7),
vmax=1,
vmin=-1,
title="Sparse inverse covariance",
)
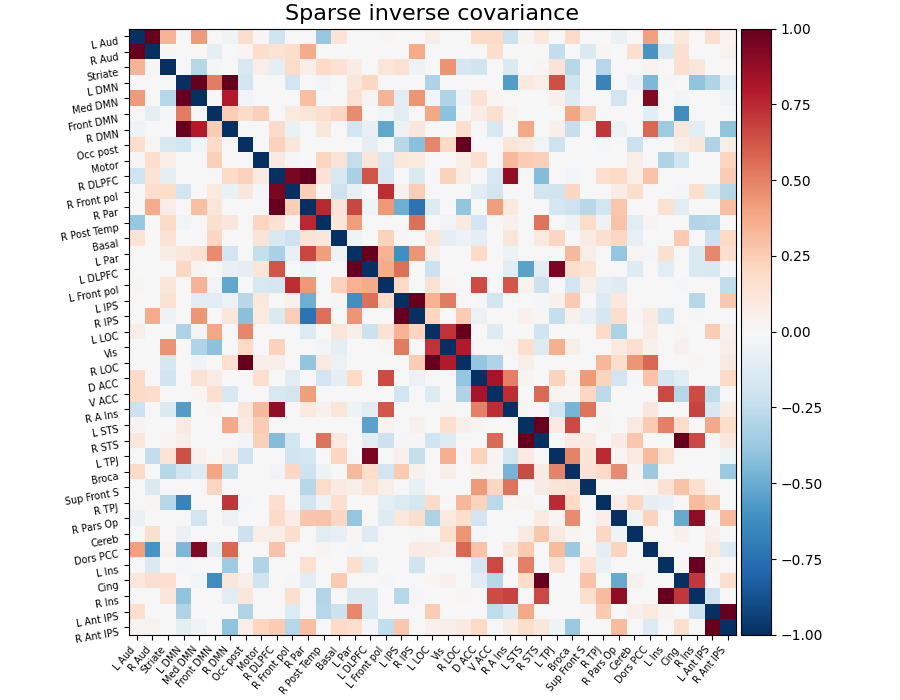
<matplotlib.image.AxesImage object at 0x7f6f3f4d9f30>
And now display the corresponding graph¶
plot_connectome(
-estimator.precision_, coords, title="Sparse inverse covariance"
)
show()
3D visualization in a web browser¶
An alternative to plot_connectome is to use
view_connectome that gives more interactive
visualizations in a web browser. See 3D Plots of connectomes
for more details.
view = view_connectome(-estimator.precision_, coords)
# In a notebook, if ``view`` is the output of a cell, it will
# be displayed below the cell
view
# uncomment this to open the plot in a web browser:
# view.open_in_browser()
Total running time of the script: (0 minutes 11.232 seconds)
Estimated memory usage: 848 MB