Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Deriving spatial maps from group fMRI data using ICA and Dictionary Learning¶
Various approaches exist to derive spatial maps or networks from group fmr data. The methods extract distributed brain regions that exhibit similar BOLD fluctuations over time. Decomposition methods allow for generation of many independent maps simultaneously without the need to provide a priori information (e.g. seeds or priors.)
This example will apply two popular decomposition methods, ICA and Dictionary learning, to fMRI data measured while children and young adults watch movies. The resulting maps will be visualized using atlas plotting tools.
CanICA is an ICA method for group-level analysis of fMRI data. Compared to other strategies, it brings a well-controlled group model, as well as a thresholding algorithm controlling for specificity and sensitivity with an explicit model of the signal.
The reference paper is Varoquaux et al.[1].
Load brain development fMRI dataset¶
from nilearn.datasets import fetch_development_fmri
rest_dataset = fetch_development_fmri(n_subjects=30)
func_filenames = rest_dataset.func # list of 4D nifti files for each subject
# print basic information on the dataset
print(f"First functional nifti image (4D) is at: {rest_dataset.func[0]}")
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
First functional nifti image (4D) is at: /home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Apply CanICA on the data¶
We use “whole-brain-template” as a strategy to compute the mask, as this leads to slightly faster and more reproducible results. However, the images need to be in MNI template space.
import warnings
from sklearn.exceptions import ConvergenceWarning
from nilearn.decomposition import CanICA
canica = CanICA(
n_components=20,
memory="nilearn_cache",
memory_level=1,
verbose=1,
random_state=0,
mask_strategy="whole-brain-template",
n_jobs=2,
)
with warnings.catch_warnings():
# silence warnings about ICA not converging
# Consider increasing tolerance or the maximum number of iterations.
warnings.filterwarnings(action="ignore", category=ConvergenceWarning)
canica.fit(func_filenames)
# Retrieve the independent components in brain space. Directly
# accessible through attribute `components_img_`.
canica_components_img = canica.components_img_
# components_img is a Nifti Image object, and can be saved to a file with
# the following lines:
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_compare_decomposition"
output_dir.mkdir(exist_ok=True, parents=True)
print(f"Output will be saved to: {output_dir}")
canica_components_img.to_filename(output_dir / "canica_resting_state.nii.gz")
[CanICA.fit] Loading data from
['/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-
pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz']
[CanICA.fit] Computing mask
[CanICA.fit] Resampling mask
[CanICA.fit] Finished fit
[CanICA.fit] Loading data
[CanICA.fit] Computing image from signals
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 10 out of 10 | elapsed: 15.1s finished
[CanICA.fit] Computing image from signals
Output will be saved to: /home/runner/work/nilearn/nilearn/examples/03_connectivity/results/plot_compare_decomposition
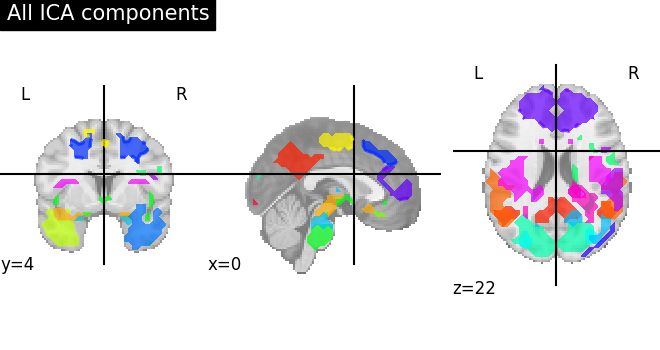
To visualize we plot the outline of all components on one figure
from nilearn.plotting import plot_prob_atlas
# Plot all ICA components together
plot_prob_atlas(canica_components_img, title="All ICA components")
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/ma/core.py:2892: UserWarning:
Warning: converting a masked element to nan.
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f32459690>
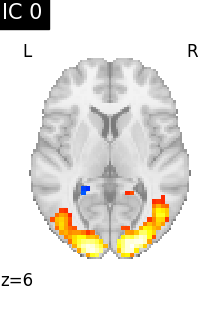
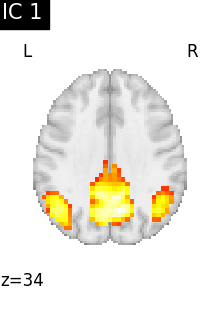
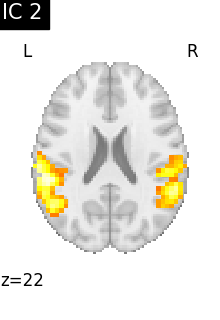
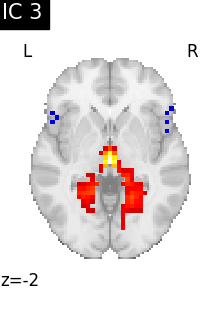
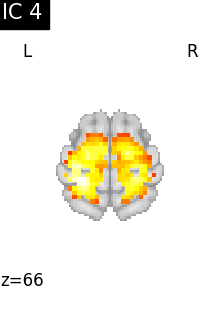
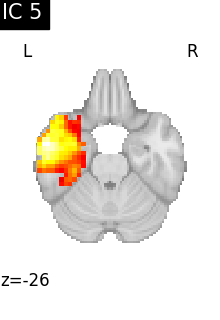
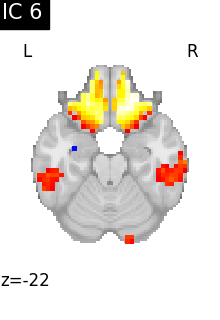
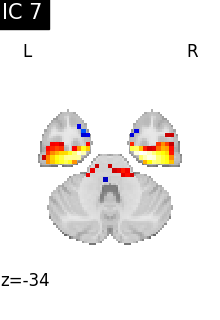
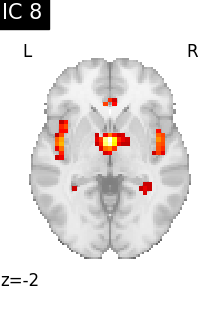
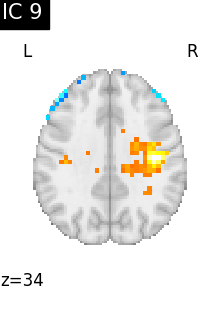
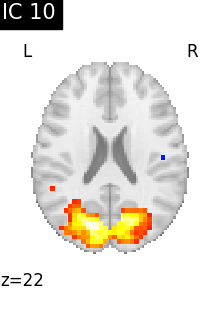
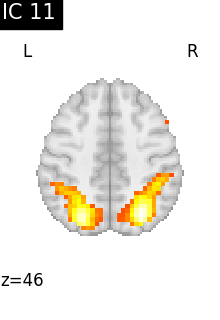
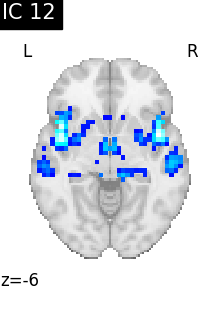
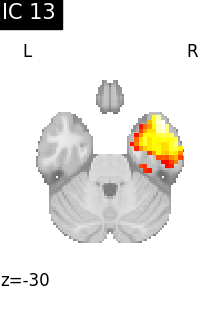
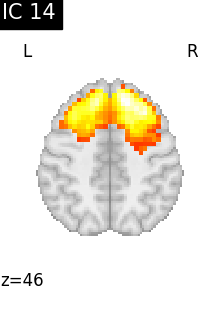
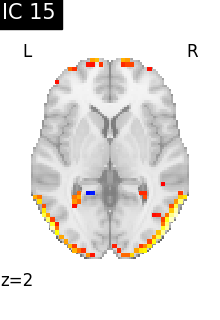
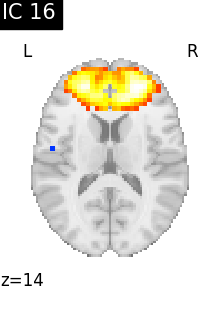
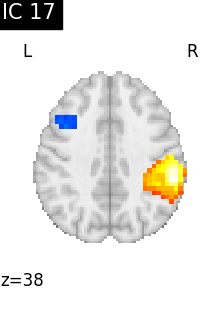
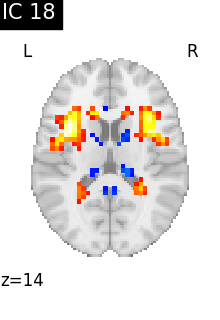
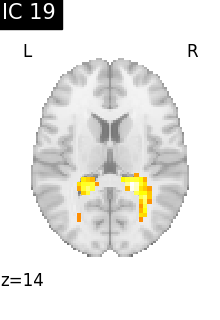
Finally, we plot the map for each ICA component separately
from nilearn.image import iter_img
from nilearn.plotting import plot_stat_map, show
for i, cur_img in enumerate(iter_img(canica_components_img)):
plot_stat_map(
cur_img,
display_mode="z",
title=f"IC {int(i)}",
cut_coords=1,
vmax=0.05,
vmin=-0.05,
colorbar=False,
)
show()
Compare CanICA to dictionary learning¶
Dictionary learning is a sparsity based decomposition method for extracting spatial maps. It extracts maps that are naturally sparse and usually cleaner than ICA. Here, we will compare networks built with CanICA to networks built with Dictionary learning.
For more detailse see Mensch et al.[2].
Create a dictionary learning estimator
from nilearn.decomposition import DictLearning
dict_learning = DictLearning(
n_components=20,
memory="nilearn_cache",
memory_level=1,
verbose=1,
random_state=0,
n_epochs=1,
mask_strategy="whole-brain-template",
n_jobs=2,
)
print("[Example] Fitting dictionary learning model")
dict_learning.fit(func_filenames)
print("[Example] Saving results")
# Grab extracted components umasked back to Nifti image.
# Note: For older versions, less than 0.4.1. components_img_
# is not implemented. See Note section above for details.
dictlearning_components_img = dict_learning.components_img_
dictlearning_components_img.to_filename(
output_dir / "dictionary_learning_resting_state.nii.gz"
)
[Example] Fitting dictionary learning model
[DictLearning.fit] Loading data from
['/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-
pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz']
[DictLearning.fit] Computing mask
[DictLearning.fit] Resampling mask
[DictLearning.fit] Finished fit
[DictLearning.fit] Loading data
[DictLearning.fit] Learning initial components
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[DictLearning.fit] Computing initial loadings
________________________________________________________________________________
[Memory] Calling nilearn.decomposition.dict_learning._compute_loadings...
_compute_loadings(array([[-0.005308, ..., -0.003948],
...,
[-0.000253, ..., 0.003836]], shape=(20, 21781)),
array([[-0.280625, ..., 0.825802],
...,
[-0.997198, ..., -0.015035]], shape=(600, 21781)))
_________________________________________________compute_loadings - 0.0s, 0.0min
[DictLearning.fit] Learning dictionary
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._dict_learning.dict_learning_online...
dict_learning_online(array([[-0.280625, ..., -0.997198],
...,
[ 0.825802, ..., -0.015035]], shape=(21781, 600)),
20, alpha=10, batch_size=20, method='cd', dict_init=array([[-0.294655, ..., -0.01288 ],
...,
[-0.29801 , ..., -0.313942]], shape=(20, 600)), verbose=0, random_state=0, return_code=True, shuffle=True, n_jobs=1, max_iter=1090)
_____________________________________________dict_learning_online - 1.0s, 0.0min
[DictLearning.fit] Computing image from signals
[Example] Saving results
Visualize the results
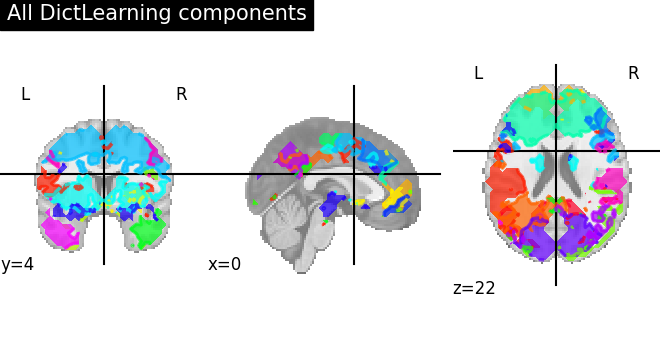
First plot all DictLearning components together
plot_prob_atlas(
dictlearning_components_img, title="All DictLearning components"
)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/ma/core.py:2892: UserWarning:
Warning: converting a masked element to nan.
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f6f4162d4b0>
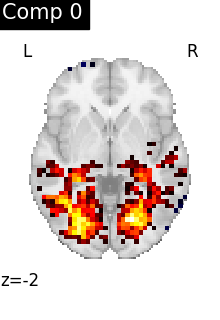
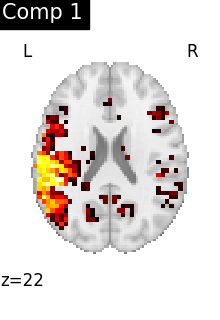
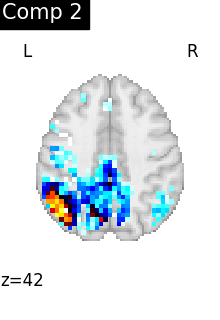
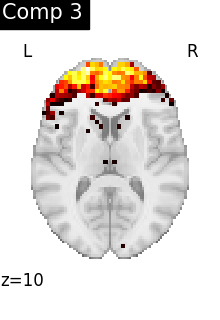
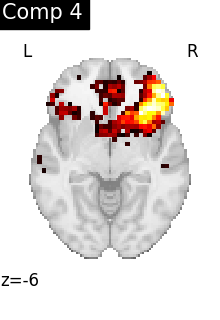
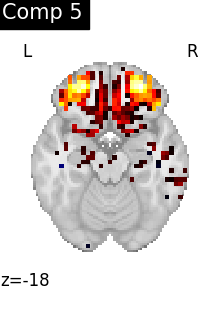
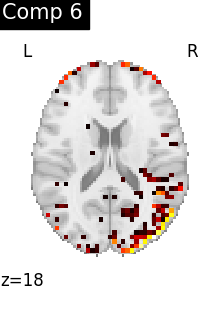
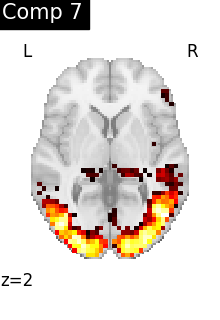
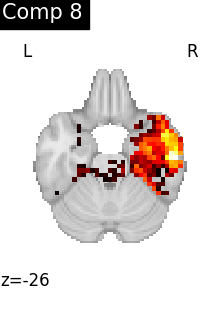
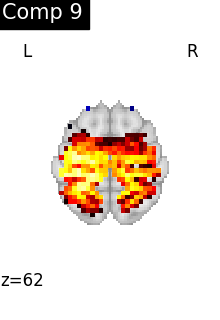
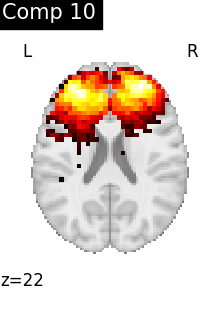
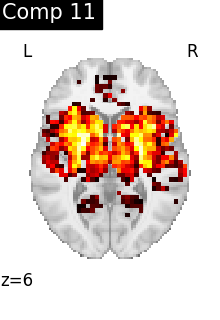
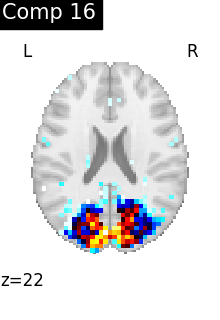
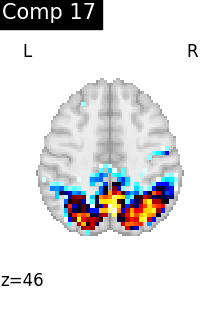
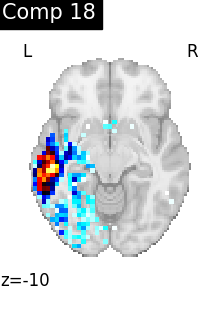
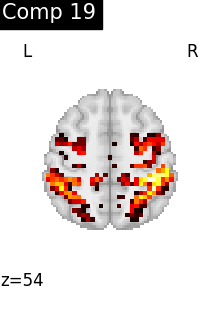
One plot of each component
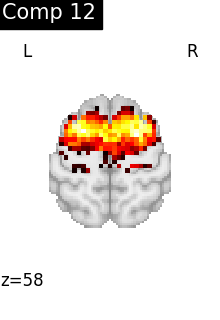
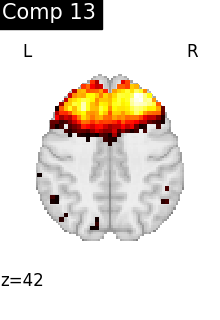
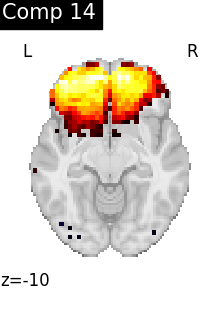
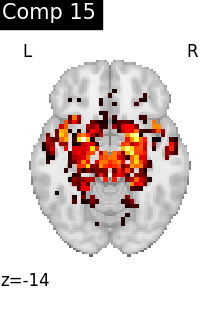
for i, cur_img in enumerate(iter_img(dictlearning_components_img)):
plot_stat_map(
cur_img,
display_mode="z",
title=f"Comp {int(i)}",
cut_coords=1,
vmax=0.1,
vmin=-0.1,
colorbar=False,
)
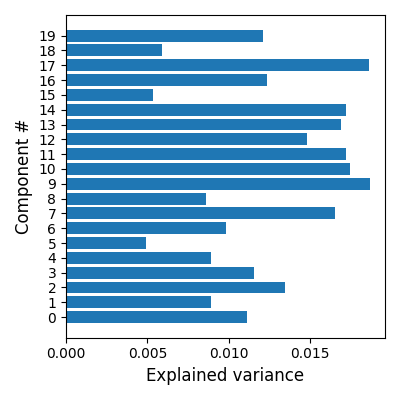
Estimate explained variance per component and plot using matplotlib
The fitted object dict_learning can be used to calculate the score per component
scores = dict_learning.score(func_filenames, per_component=True)
# Plot the scores
import numpy as np
from matplotlib import pyplot as plt
from matplotlib.ticker import FormatStrFormatter
plt.figure(figsize=(4, 4), constrained_layout=True)
positions = np.arange(len(scores))
plt.barh(positions, scores)
plt.ylabel("Component #", size=12)
plt.xlabel("Explained variance", size=12)
plt.yticks(np.arange(20))
plt.gca().xaxis.set_major_formatter(FormatStrFormatter("%.3f"))
show()
________________________________________________________________________________
[Memory] Calling nilearn.decomposition._base._explained_variance...
_explained_variance(array([[-2.806378e-01, ..., 8.257976e-01],
...,
[-2.074250e-15, ..., -4.247761e-16]], shape=(5040, 21781)),
array([[0. , ..., 0.002218],
...,
[0. , ..., 0. ]], shape=(20, 21781)), per_component=True)
______________________________________________explained_variance - 14.0s, 0.2min
Note
To see how to extract subject-level timeseries’ from regions created using Dictionary learning, see example Regions extraction using dictionary learning and functional connectomes.
References¶
Total running time of the script: (1 minutes 25.368 seconds)
Estimated memory usage: 2621 MB