Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Working with long time series fMRI images¶
In this example, we will demonstrate how one can work with large fMRI images
more efficiently. Note that fMRI images can be large on-disk due to several
different factors, including a long acquisition or high-resolution sampling.
Currently, this example focuses on memory-efficient interactions with long
time series fMRI data. In this case, loading the whole time series into memory
may represent a significant computational cost. We will therefore explore
strategies to minimize the amount of data that is loaded into memory,
and we will compare these strategies against a naive usage of
NiftiMasker.
To make this more concrete, we will create a large fMRI image with over
800 time points by concatenating individual subjects in the
fetch_adhd dataset.
Our goal is to extract data from several regions of interest (ROIs)
defined by a number of binary masks, all in parallel.
When using NiftiMasker to extract data from each
ROI in parallel, each parallel process will load the entire fMRI image into
memory. This can lead to a significant increase in memory usage and may be
infeasible in some computational environments.
We will thus compare three different methods for this task, each handling the fMRI image in a different way:
A naive, unoptimized usage of
NiftiMasker.Masking the fMRI image’s data array using numpy indexing.
For the first method, there are two ways to input the fMRI image:
passing the file path (i.e., the location of the large fMRI image on-disk).
loading image first and passing this in-memory object to joblib.
When using file paths, the entire image is loaded into memory for each process, and that is exactly the problem we described earlier.
However, when the fMRI image is loaded once and then passed to
joblib.Parallel as an in-memory object, the image is not loaded
multiple times. We will see that this can already be a significant
improvement over the naive usage of NiftiMasker with
file paths.
Create a large fMRI image¶
Here we will create a “large” fMRI image by fetching 6 subjects’
fMRI images via the fetch_adhd
function, concatenating them and then saving to a file.
from pathlib import Path
from nilearn.datasets import fetch_adhd
from nilearn.image import concat_imgs
N_SUBJECTS = 6
N_REGIONS = 2
def create_large_fmri(n_subjects):
fmri_data = fetch_adhd(n_subjects=n_subjects)
fmri_img = concat_imgs(fmri_data.func)
n_timepoints = fmri_img.shape[-1]
output_dir = Path.cwd() / "results" / "plot_mask_large_fmri"
output_dir.mkdir(parents=True, exist_ok=True)
print(f"Large fmri file will be saved to:\n{output_dir}")
fmri_path = output_dir / "large_fmri.nii.gz"
fmri_img.to_filename(fmri_path)
return fmri_path, n_timepoints
fmri_path, n_timepoints = create_large_fmri(N_SUBJECTS)
[fetch_adhd] Dataset found in /home/runner/nilearn_data/adhd
[fetch_adhd] Downloading data from
https://www.nitrc.org/frs/download.php/7784/adhd40_0010128.tgz ...
[fetch_adhd] ...done. (1 seconds, 0 min)
[fetch_adhd] Extracting data from
/home/runner/nilearn_data/adhd/67b924beca01574bbea08fde49fbbbcd/adhd40_0010128.t
gz...
[fetch_adhd] .. done.
[fetch_adhd] Downloading data from
https://www.nitrc.org/frs/download.php/7785/adhd40_0021019.tgz ...
[fetch_adhd] ...done. (1 seconds, 0 min)
[fetch_adhd] Extracting data from
/home/runner/nilearn_data/adhd/67b924beca01574bbea08fde49fbbbcd/adhd40_0021019.t
gz...
[fetch_adhd] .. done.
[fetch_adhd] Downloading data from
https://www.nitrc.org/frs/download.php/7786/adhd40_0023008.tgz ...
[fetch_adhd] ...done. (0 seconds, 0 min)
[fetch_adhd] Extracting data from
/home/runner/nilearn_data/adhd/67b924beca01574bbea08fde49fbbbcd/adhd40_0023008.t
gz...
[fetch_adhd] .. done.
[fetch_adhd] Downloading data from
https://www.nitrc.org/frs/download.php/7787/adhd40_0023012.tgz ...
[fetch_adhd] ...done. (0 seconds, 0 min)
[fetch_adhd] Extracting data from
/home/runner/nilearn_data/adhd/67b924beca01574bbea08fde49fbbbcd/adhd40_0023012.t
gz...
[fetch_adhd] .. done.
Large fmri file will be saved to:
/home/runner/work/nilearn/nilearn/examples/07_advanced/results/plot_mask_large_fmri
Create a set of binary masks¶
We will now create 4 binary masks from a brain atlas. Here we will use the
multiscale functional brain parcellations via the
fetch_atlas_basc_multiscale_2015 function.
We will fetch a 64-region version of this atlas and then create separate
binary masks for the first 4 regions.
In this case, the atlas and the fMRI image have different resolutions.
So we will resample the atlas to the fMRI image. It is important to do that
because only NiftiMasker can handle the resampling
of the mask to the fMRI image but other methods considered here will not.
from nilearn.datasets import fetch_atlas_basc_multiscale_2015
from nilearn.image import load_img, new_img_like, resample_to_img
def create_masks(fmri_path, n_regions):
atlas_path = fetch_atlas_basc_multiscale_2015(resolution=64).maps
atlas_img = load_img(atlas_path)
resampled_atlas = resample_to_img(
atlas_img, fmri_path, interpolation="nearest"
)
mask_paths = []
output_dir = Path.cwd() / "results" / "plot_mask_large_fmri"
for idx in range(1, n_regions + 1):
mask = resampled_atlas.get_fdata() == idx
mask = new_img_like(
ref_niimg=fmri_path, data=mask, affine=resampled_atlas.affine
)
path = output_dir / f"mask_{idx}.nii.gz"
mask.to_filename(path)
mask_paths.append(path)
return mask_paths
mask_paths = create_masks(fmri_path, N_REGIONS)
[fetch_atlas_basc_multiscale_2015] Dataset found in
/home/runner/nilearn_data/basc_multiscale_2015
Mask the fMRI image using NiftiMasker¶
Let’s first mask the fMRI image using NiftiMasker.
This is the most user-friendly way to extract data from an fMRI image as it
makes it easy to standardize, smooth, detrend, etc. the data.
We will first wrap the nilearn.maskers.NiftiMasker.fit_transform
within a function so that it is more readable and easier to use.
We will then define another function that would mask the fMRI image using
multiple masks in parallel using the joblib package. As mentioned
earlier, this could further be done in two ways: directly using file paths
as input or first loading the images in-memory and passing them as input.
So we will define two functions for each case.
We can then track the memory usage of these functions via the
memory_profiler package.
from joblib import Parallel, delayed
from nilearn.maskers import NiftiMasker
def nifti_masker_single(fmri, mask):
return NiftiMasker(mask_img=mask, verbose=1).fit_transform(fmri)
def nifti_masker_parallel_path(fmri_path, mask_paths):
Parallel(n_jobs=N_REGIONS)(
delayed(nifti_masker_single)(fmri_path, mask_path)
for mask_path in mask_paths
)
def nifti_masker_parallel_inmemory(fmri_path, mask_paths):
fmri_img = load_img(fmri_path)
mask_imgs = [load_img(mask) for mask in mask_paths]
Parallel(n_jobs=N_REGIONS)(
delayed(nifti_masker_single)(fmri_img, mask) for mask in mask_imgs
)
Let’s also create a dictionary to store the memory usage for each method.
from memory_profiler import memory_usage
nifti_masker = {"path": [], "in_memory": []}
nifti_masker["path"] = memory_usage(
(nifti_masker_parallel_path, (fmri_path, mask_paths)),
max_usage=True,
include_children=True,
multiprocess=True,
)
nifti_masker["in_memory"] = memory_usage(
(nifti_masker_parallel_inmemory, (fmri_path, mask_paths)),
max_usage=True,
include_children=True,
multiprocess=True,
)
print(
f"Peak memory usage with NiftiMasker, {N_REGIONS} jobs in parallel:\n"
f"- with file paths: {nifti_masker['path']} MiB\n"
f"- with in-memory images: {nifti_masker['in_memory']} MiB"
)
Peak memory usage with NiftiMasker, 2 jobs in parallel:
- with file paths: 10816.66015625 MiB
- with in-memory images: 10899.390625 MiB
Masking using numpy indexing¶
Now let’s see how we can mask the fMRI image using numpy indexing. This could be more efficient than the NiftiMasker when we simply need to mask the fMRI image with binary masks and don’t need to standardize, smooth, etc. the image.
As before we will first define a function that would mask the data array of the fMRI image using a single mask. We will then define another function that would iterate over multiple masks in parallel and mask the data array of the fMRI image using each mask.
import numpy as np
def numpy_masker_single(fmri, mask):
return fmri[mask]
def numpy_masker_parallel(fmri_path, mask_paths):
fmri_data = load_img(fmri_path).get_fdata()
masks = [load_img(mask).get_fdata().astype(bool) for mask in mask_paths]
Parallel(n_jobs=N_REGIONS)(
delayed(numpy_masker_single)(fmri_data, mask) for mask in masks
)
Let’s measure the memory usage
numpy_masker = memory_usage(
(numpy_masker_parallel, (fmri_path, mask_paths)),
max_usage=True,
include_children=True,
multiprocess=True,
)
print(
f"Peak memory usage with numpy indexing, {N_REGIONS} jobs in parallel:\n"
f"{numpy_masker} MiB"
)
Peak memory usage with numpy indexing, 2 jobs in parallel:
9742.5546875 MiB
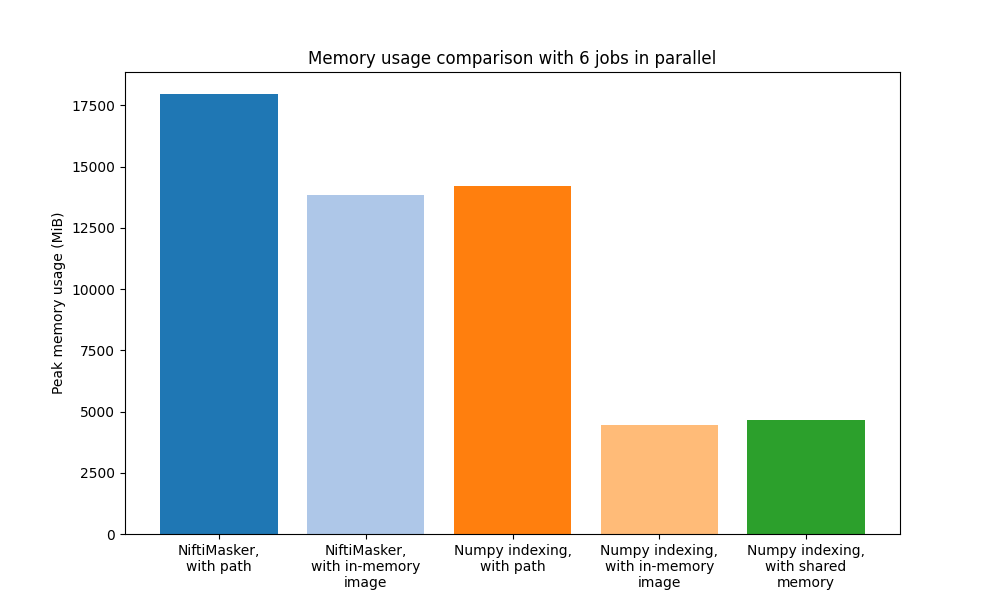
Conclusion¶
So overall we can see that there are much more memory-efficient ways to
extract ROIs in parallel from large fMRI images than simply
using NiftiMasker with file paths.
The two methods that standout both use numpy indexing and involve
loading the fMRI image prior to masking. In fact, loading the fMRI image
prior is generally better than using file paths even with
NiftiMasker.
So if your goal is to simply extract data from an fMRI image, numpy indexing
is much more memory-efficient than using
NiftiMasker.
However, if you also need to standardize, smooth, detrend, etc. the data,
then using NiftiMasker with in-memory images is
the most user-friendly way to run all these operations in the appropriate
order while still being relatively memory-efficient.
Finally, it should be noted that the differences in memory usage between
the methods can be more significant when working with even larger images
and/or more jobs/regions in parallel. You can try increasing the
N_SUBJECTS and N_REGIONS variables at the beginning of this example
to see how the memory usage changes for each method.
Total running time of the script: (0 minutes 37.897 seconds)
Estimated memory usage: 4498 MB