Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.plotting.find_parcellation_cut_coords¶
- nilearn.plotting.find_parcellation_cut_coords(labels_img, background_label=0, return_label_names=False, label_hemisphere='left')[source]¶
Return coordinates of center of mass of 3D parcellation atlas.
- Parameters:
- labels_img3D Nifti1Image
A brain parcellation atlas with specific mask labels for each parcellated region.
- background_label
int, default=0 Label value used in labels_img to represent background.
- return_label_names
bool, default=False Returns list of labels.
- label_hemisphere‘left’ or ‘right’, default=’left’
Choice of hemisphere to compute label center coords for. Applies only in cases where atlas labels are lateralized. Eg. Yeo or Harvard Oxford atlas.
- Returns:
- coordsnumpy.ndarray of shape (n_labels, 3)
Label regions cut coordinates in image space (mm).
- labels_list
list, optional Label region. Returned only when return_label_names is True.
See also
nilearn.plotting.find_probabilistic_atlas_cut_coordsFor coordinates extraction on probabilistic atlases (4D) (Eg. MSDL atlas)
Examples using nilearn.plotting.find_parcellation_cut_coords¶
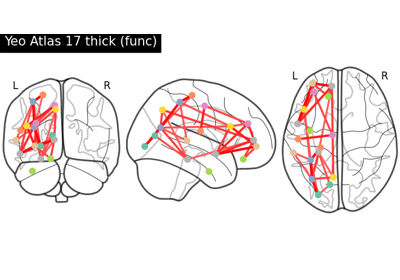
Comparing connectomes on different reference atlases