Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Different classifiers in decoding the Haxby dataset¶
Here we compare different classifiers on a visual object recognition decoding task.
# We start by loading data using nilearn dataset fetcher
from nilearn import datasets
from nilearn.image import get_data
from nilearn.plotting import plot_stat_map, show
Loading the data¶
# by default 2nd subject data will be fetched
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print(
"First subject anatomical nifti image (3D) located is "
f"at: {haxby_dataset.anat[0]}"
)
print(
"First subject functional nifti image (4D) is located "
f"at: {haxby_dataset.func[0]}"
)
# load labels
import numpy as np
import pandas as pd
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
stimuli = labels["labels"]
# identify resting state (baseline) labels in order to be able to remove them
resting_state = stimuli == "rest"
# extract the indices of the images corresponding to some condition or task
task_mask = np.logical_not(resting_state)
# find names of remaining active labels
categories = stimuli[task_mask].unique()
# extract tags indicating to which acquisition run a tag belongs
run_labels = labels["chunks"][task_mask]
# Load the fMRI data
# For decoding, standardizing is often very important
mask_filename = haxby_dataset.mask_vt[0]
func_filename = haxby_dataset.func[0]
# Because the data is in one single large 4D image, we need to use
# index_img to do the split easily.
from nilearn.image import index_img
fmri_niimgs = index_img(func_filename, task_mask)
classification_target = stimuli[task_mask]
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
First subject anatomical nifti image (3D) located is at: /home/runner/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Training the decoder¶
# Then we define the various classifiers that we use
classifiers = [
"svc_l2",
"svc_l1",
"logistic_l1",
"logistic_l2",
"ridge_classifier",
]
# Here we compute prediction scores and run time for all these
# classifiers
import time
from sklearn.model_selection import LeaveOneGroupOut
from nilearn.decoding import Decoder
cv = LeaveOneGroupOut()
classifiers_data = {}
for classifier_name in sorted(classifiers):
print(70 * "_")
# The decoder has as default score the `roc_auc`
decoder = Decoder(
estimator=classifier_name,
mask=mask_filename,
cv=cv,
screening_percentile=100,
verbose=1,
)
t0 = time.time()
decoder.fit(
fmri_niimgs,
classification_target,
groups=run_labels,
)
classifiers_data[classifier_name] = {"score": decoder.cv_scores_}
print(f"{classifier_name:10}: {time.time() - t0:.2f}s")
for category in categories:
mean = np.mean(classifiers_data[classifier_name]["score"][category])
std = np.std(classifiers_data[classifier_name]["score"][category])
print(f" {category:14} vs all -- AUC: {mean:1.2f} +- {std:1.2f}")
# Adding the average performance per estimator
scores = classifiers_data[classifier_name]["score"]
scores["AVERAGE"] = np.mean(list(scores.values()), axis=0)
classifiers_data[classifier_name]["score"] = scores
______________________________________________________________________
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:101: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
[Parallel(n_jobs=1)]: Done 40 tasks | elapsed: 1.5min
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
[Parallel(n_jobs=1)]: Done 96 out of 96 | elapsed: 3.4min finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
logistic_l1: 204.14s
scissors vs all -- AUC: 0.90 +- 0.08
face vs all -- AUC: 0.97 +- 0.04
cat vs all -- AUC: 0.97 +- 0.04
shoe vs all -- AUC: 0.91 +- 0.08
house vs all -- AUC: 1.00 +- 0.00
scrambledpix vs all -- AUC: 0.97 +- 0.08
bottle vs all -- AUC: 0.82 +- 0.17
chair vs all -- AUC: 0.87 +- 0.17
______________________________________________________________________
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:101: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
[Parallel(n_jobs=1)]: Done 40 tasks | elapsed: 1.5min
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
[Parallel(n_jobs=1)]: Done 96 out of 96 | elapsed: 3.4min finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
logistic_l2: 204.10s
scissors vs all -- AUC: 0.90 +- 0.08
face vs all -- AUC: 0.97 +- 0.04
cat vs all -- AUC: 0.97 +- 0.04
shoe vs all -- AUC: 0.91 +- 0.08
house vs all -- AUC: 1.00 +- 0.00
scrambledpix vs all -- AUC: 0.97 +- 0.08
bottle vs all -- AUC: 0.82 +- 0.17
chair vs all -- AUC: 0.87 +- 0.17
______________________________________________________________________
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:101: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 40 tasks | elapsed: 3.1s
[Parallel(n_jobs=1)]: Done 96 out of 96 | elapsed: 7.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
ridge_classifier: 9.49s
scissors vs all -- AUC: 0.94 +- 0.05
face vs all -- AUC: 0.98 +- 0.02
cat vs all -- AUC: 0.95 +- 0.04
shoe vs all -- AUC: 0.94 +- 0.07
house vs all -- AUC: 1.00 +- 0.00
scrambledpix vs all -- AUC: 1.00 +- 0.00
bottle vs all -- AUC: 0.89 +- 0.09
chair vs all -- AUC: 0.93 +- 0.07
______________________________________________________________________
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:101: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 40 tasks | elapsed: 9.0s
[Parallel(n_jobs=1)]: Done 96 out of 96 | elapsed: 17.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
svc_l1 : 19.59s
scissors vs all -- AUC: 0.92 +- 0.05
face vs all -- AUC: 0.98 +- 0.03
cat vs all -- AUC: 0.96 +- 0.04
shoe vs all -- AUC: 0.92 +- 0.07
house vs all -- AUC: 1.00 +- 0.00
scrambledpix vs all -- AUC: 0.99 +- 0.01
bottle vs all -- AUC: 0.89 +- 0.08
chair vs all -- AUC: 0.93 +- 0.04
______________________________________________________________________
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:101: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f4164c6a0>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 40 tasks | elapsed: 24.1s
[Parallel(n_jobs=1)]: Done 96 out of 96 | elapsed: 50.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
svc_l2 : 52.18s
scissors vs all -- AUC: 0.90 +- 0.09
face vs all -- AUC: 0.96 +- 0.05
cat vs all -- AUC: 0.96 +- 0.04
shoe vs all -- AUC: 0.91 +- 0.08
house vs all -- AUC: 1.00 +- 0.00
scrambledpix vs all -- AUC: 0.96 +- 0.10
bottle vs all -- AUC: 0.82 +- 0.17
chair vs all -- AUC: 0.87 +- 0.16
Visualization¶
# Then we make a rudimentary diagram
import matplotlib.pyplot as plt
plt.subplots(figsize=(8, 6), constrained_layout=True)
all_categories = np.sort(np.hstack([categories, "AVERAGE"]))
tick_position = np.arange(len(all_categories))
plt.yticks(tick_position + 0.25, all_categories)
height = 0.1
for color, classifier_name in zip(
["b", "m", "k", "r", "g"], classifiers, strict=False
):
score_means = [
np.mean(classifiers_data[classifier_name]["score"][category])
for category in all_categories
]
plt.barh(
tick_position,
score_means,
label=classifier_name.replace("_", " "),
height=height,
color=color,
)
tick_position = tick_position + height
plt.xlabel("Classification accuracy (AUC score)")
plt.ylabel("Visual stimuli category")
plt.xlim(xmin=0.5)
plt.legend(ncol=1, bbox_to_anchor=(1.3, 0.2))
plt.title(
"Category-specific classification accuracy for different classifiers"
)
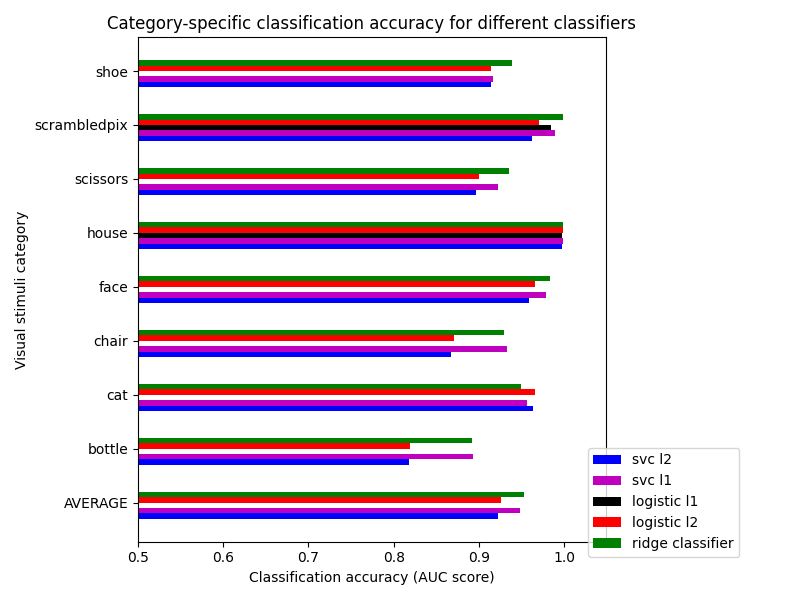
Text(0.5, 1.0, 'Category-specific classification accuracy for different classifiers')
We can see that for a fixed penalty the results are similar between the svc and the logistic regression. The main difference relies on the penalty l1and l2). The sparse penalty works better because we are in an intra-subject setting.
Visualizing the face vs house map¶
Restrict the decoding to face vs house
condition_mask = np.logical_or(stimuli == "face", stimuli == "house")
stimuli = stimuli[condition_mask]
assert len(stimuli) == 216
fmri_niimgs_condition = index_img(func_filename, condition_mask)
run_labels = labels["chunks"][condition_mask]
categories = stimuli.unique()
assert len(categories) == 2
for classifier_name in sorted(classifiers):
decoder = Decoder(
estimator=classifier_name,
mask=mask_filename,
cv=cv,
screening_percentile=100,
)
decoder.fit(fmri_niimgs_condition, stimuli, groups=run_labels)
classifiers_data[classifier_name] = {}
classifiers_data[classifier_name]["score"] = decoder.cv_scores_
classifiers_data[classifier_name]["map"] = decoder.coef_img_["face"]
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:185: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:185: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/linear_model/_logistic.py:1901: UserWarning:
l1_ratios parameter is only used when penalty is 'elasticnet'. Got (penalty=l2)
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:185: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:185: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_different_estimators.py:185: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
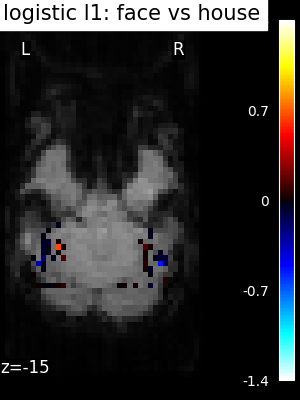
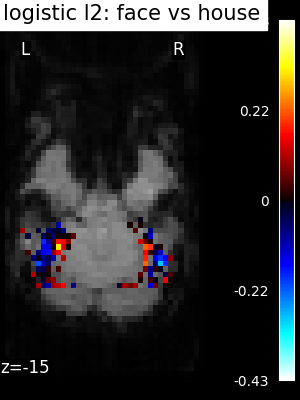
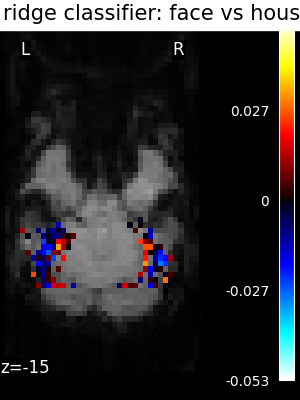
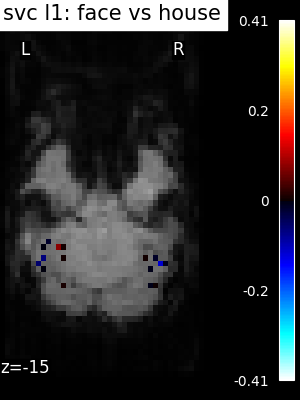
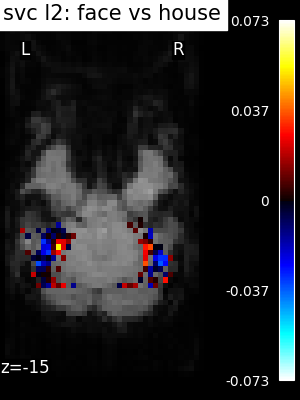
Finally, we plot the face vs house map for the different classifiers Use the average EPI as a background
from nilearn.image import mean_img
mean_epi_img = mean_img(func_filename)
for classifier_name in sorted(classifiers):
coef_img = classifiers_data[classifier_name]["map"]
threshold = np.max(np.abs(get_data(coef_img))) * 1e-3
plot_stat_map(
coef_img,
bg_img=mean_epi_img,
display_mode="z",
cut_coords=[-15],
threshold=threshold,
title=f"{classifier_name.replace('_', ' ')}: face vs house",
figure=plt.figure(figsize=(3, 4)),
)
show()
Total running time of the script: (8 minutes 30.631 seconds)
Estimated memory usage: 1329 MB