Note
Go to the end to download the full example code or to run this example in your browser via Binder.
ROI-based decoding analysis in Haxby et al. dataset¶
In this script we reproduce the data analysis conducted by Haxby et al.[1].
Specifically, we look at decoding accuracy for different objects in three different masks: the full ventral stream (mask_vt), the house selective areas (mask_house) and the face selective areas (mask_face), that have been defined via a standard GLM-based analysis.
# Fetch data using nilearn dataset fetcher
from nilearn import datasets
from nilearn.plotting import show
Load and prepare the data¶
# by default we fetch 2nd subject data for analysis
haxby_dataset = datasets.fetch_haxby()
func_filename = haxby_dataset.func[0]
# Print basic information on the dataset
print(
"First subject anatomical nifti image (3D) located is "
f"at: {haxby_dataset.anat[0]}"
)
print(
f"First subject functional nifti image (4D) is located at: {func_filename}"
)
# load labels
import pandas as pd
# Load nilearn NiftiMasker, the practical masking and unmasking tool
from nilearn.maskers import NiftiMasker
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
stimuli = labels["labels"]
# identify resting state labels in order to be able to remove them
task_mask = stimuli != "rest"
# find names of remaining active labels
categories = stimuli[task_mask].unique()
# extract tags indicating to which acquisition run a tag belongs
run_labels = labels["chunks"][task_mask]
# apply the task_mask to fMRI data (func_filename)
from nilearn.image import index_img
task_data = index_img(func_filename, task_mask)
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
First subject anatomical nifti image (3D) located is at: /home/runner/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Decoding on the different masks¶
The classifier used here is a support vector classifier (svc).
We use
Decoder and specify the classifier.
import numpy as np
# Make a data splitting object for cross validation
from sklearn.model_selection import LeaveOneGroupOut
from nilearn.decoding import Decoder
cv = LeaveOneGroupOut()
We use Decoder to estimate a baseline.
import warnings
mask_names = ["mask_vt", "mask_face", "mask_house"]
mask_scores = {}
mask_chance_scores = {}
for mask_name in mask_names:
print(f"Working on {mask_name}")
# For decoding, standardizing is often very important
mask_filename = haxby_dataset[mask_name][0]
masker = NiftiMasker(mask_img=mask_filename, verbose=1)
mask_scores[mask_name] = {}
mask_chance_scores[mask_name] = {}
for category in categories:
print(f"Processing {mask_name} {category}")
classification_target = stimuli[task_mask] == category
# Specify the classifier to the decoder object.
# With the decoder we can input the masker directly.
# We are using the svc_l1 here because it is intra subject.
decoder = Decoder(
estimator="svc_l1",
cv=cv,
mask=masker,
scoring="roc_auc",
verbose=1,
)
with warnings.catch_warnings():
# ignore warnings thrown because the ROI mask we are using
# are much smaller than the whole brain.
warnings.filterwarnings(action="ignore", category=UserWarning)
decoder.fit(task_data, classification_target, groups=run_labels)
mask_scores[mask_name][category] = decoder.cv_scores_[1]
mean = np.mean(mask_scores[mask_name][category])
std = np.std(mask_scores[mask_name][category])
print(f"Scores: {mean:1.2f} +- {std:1.2f}")
dummy_classifier = Decoder(
estimator="dummy_classifier",
cv=cv,
mask=masker,
scoring="roc_auc",
verbose=1,
)
with warnings.catch_warnings():
# ignore warnings thrown because the ROI mask we are using
# are much smaller than the whole brain.
warnings.filterwarnings(action="ignore", category=UserWarning)
dummy_classifier.fit(
task_data, classification_target, groups=run_labels
)
mask_chance_scores[mask_name][category] = dummy_classifier.cv_scores_[
1
]
Working on mask_vt
Processing mask_vt scissors
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.70 +- 0.22
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt face
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.8s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.68 +- 0.24
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt cat
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.70 +- 0.18
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt shoe
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.66 +- 0.17
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt house
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.8s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.66 +- 0.19
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt scrambledpix
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.77 +- 0.21
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt bottle
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.74 +- 0.21
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Processing mask_vt chair
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.9s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.60 +- 0.32
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 22837.5mm^3 = 22.8375cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 464 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.1s finished
Working on mask_face
Processing mask_face scissors
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.2s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.72 +- 0.14
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face face
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.55 +- 0.18
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face cat
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.57 +- 0.11
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face shoe
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.65 +- 0.16
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face house
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.67 +- 0.10
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face scrambledpix
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.80 +- 0.09
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face bottle
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.76 +- 0.11
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_face chair
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.58 +- 0.16
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_face_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1476.56mm^3 = 1.47656cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Working on mask_house
Processing mask_house scissors
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.69 +- 0.17
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house face
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.2s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.64 +- 0.27
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house cat
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.72 +- 0.14
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house shoe
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.63 +- 0.18
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house house
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.34 +- 0.17
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house scrambledpix
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.69 +- 0.17
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house bottle
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.77 +- 0.16
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
Processing mask_house chair
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Scores: 0.73 +- 0.13
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask8b_house_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f6f41421c30>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 5807.81mm^3 = 5.80781cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 20
[Decoder.fit] Corrected screening-percentile: 100
[Decoder.fit] The decoding model will be trained on 118 features.
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 0.0s finished
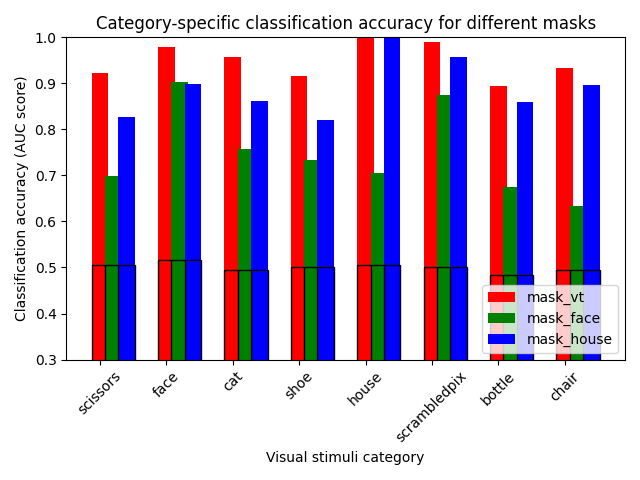
We make a simple bar plot to summarize the results¶
import matplotlib.pyplot as plt
plt.figure(constrained_layout=True)
tick_position = np.arange(len(categories))
plt.xticks(tick_position, categories, rotation=45)
for color, mask_name in zip("rgb", mask_names, strict=False):
score_means = [
np.mean(mask_scores[mask_name][category]) for category in categories
]
plt.bar(
tick_position, score_means, label=mask_name, width=0.25, color=color
)
score_chance = [
np.mean(mask_chance_scores[mask_name][category])
for category in categories
]
plt.bar(
tick_position,
score_chance,
width=0.25,
edgecolor="k",
facecolor="none",
)
tick_position = tick_position + 0.2
plt.ylabel("Classification accuracy (AUC score)")
plt.xlabel("Visual stimuli category")
plt.ylim(0.3, 1)
plt.legend(loc="lower right")
plt.title("Category-specific classification accuracy for different masks")
show()
References¶
Total running time of the script: (1 minutes 39.857 seconds)
Estimated memory usage: 1322 MB