Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Classification of age groups using functional connectivity¶
This example compares different kinds of functional connectivity between regions of interest : correlation, partial correlation, and tangent space embedding.
The resulting connectivity coefficients can be used to discriminate children from adults.In general, the tangent space embedding outperforms the standard correlations: see Dadi et al.[1] for a careful study.
Load brain development fMRI dataset and MSDL atlas¶
We study only 30 subjects from the dataset, to save computation time.
from nilearn.datasets import fetch_atlas_msdl, fetch_development_fmri
from nilearn.plotting import plot_connectome, plot_matrix, show
development_dataset = fetch_development_fmri(n_subjects=30)
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3ea4712b400183b70b7/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3eb2286e80019c3c194/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e52286e80018c3e439/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e72286e80017c41b3d/ ...
[fetch_development_fmri] ...done. (3 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e4a743a9001760814f/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e54712b400183b70a5/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e14712b400183b7097/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e32286e80018c3e42c/ ...
[fetch_development_fmri] ...done. (3 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e9a743a90017608158/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3e82286e80018c3e443/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb47052f2be3c0017057069/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e5c353c5800199ac79f/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb47045a3bc970019f073a0/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e913992690018133b1c/ ...
[fetch_development_fmri] ...done. (3 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3912286e80018c3e393/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3952286e80017c41a1b/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb47023353c58001c9ac02b/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46eaa39926900160f69af/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff391a743a900176080a9/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3914712b400173b5329/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb4702a353c58001b9cb5ae/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e9b39926900190fad5c/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38f2286e80018c3e38d/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3914712b4001a3b5579/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb470413992690018133d8c/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e9a353c58001c9abeac/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb47016a3bc970017efe44f/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e43f2be3c0017056b8a/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff37da743a90018606df1/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff37c2286e80019c3c102/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb47056353c58001c9ac064/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e5af2be3c001801f799/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38ca743a90018606dfe/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38ca743a9001760809e/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff389a743a9001660a016/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38c2286e80016c3c2da/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3884712b400183b7023/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3884712b400193b5b5c/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3872286e80017c419ea/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3872286e80017c419e9/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb4702f39926900171090ee/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e8b353c58001c9abe98/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3842286e80017c419e0/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3854712b4001a3b5568/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3814712b4001a3b5561/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3832286e80016c3c2d1/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3822286e80018c3e37b/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff382a743a90018606df8/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff37e2286e80016c3c2cb/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff3832286e80019c3c10f/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff37d4712b400193b5b54/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff37d4712b400183b7011/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb4701e3992690018133d4f/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e6b353c58001b9cb34f/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb4703bf2be3c001801fa49/ ...
[fetch_development_fmri] ...done. (2 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5cb46e92a3bc970019f0717f/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38c4712b4001a3b5573/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
[fetch_development_fmri] Downloading data from
https://osf.io/download/5c8ff38da743a900176080a2/ ...
[fetch_development_fmri] ...done. (1 seconds, 0 min)
We use probabilistic regions of interest (ROIs) from the MSDL atlas.
msdl_data = fetch_atlas_msdl()
msdl_coords = msdl_data.region_coords
n_regions = len(msdl_coords)
print(
f"MSDL has {n_regions} ROIs, "
f"part of the following networks:\n{msdl_data.networks}."
)
[fetch_atlas_msdl] Dataset found in /home/runner/nilearn_data/msdl_atlas
MSDL has 39 ROIs, part of the following networks:
['Aud', 'Aud', 'Striate', 'DMN', 'DMN', 'DMN', 'DMN', 'Occ post', 'Motor', 'R V Att', 'R V Att', 'R V Att', 'R V Att', 'Basal', 'L V Att', 'L V Att', 'L V Att', 'D Att', 'D Att', 'Vis Sec', 'Vis Sec', 'Vis Sec', 'Salience', 'Salience', 'Salience', 'Temporal', 'Temporal', 'Language', 'Language', 'Language', 'Language', 'Language', 'Cereb', 'Dors PCC', 'Cing-Ins', 'Cing-Ins', 'Cing-Ins', 'Ant IPS', 'Ant IPS'].
Region signals extraction¶
To extract regions time series, we instantiate a
NiftiMapsMasker object and pass the atlas the
file name to it, as well as filtering band-width and detrending option.
from nilearn.maskers import NiftiMapsMasker
masker = NiftiMapsMasker(
msdl_data.maps,
resampling_target="data",
t_r=development_dataset.t_r,
detrend=True,
low_pass=0.1,
high_pass=0.01,
memory="nilearn_cache",
memory_level=1,
standardize_confounds=True,
verbose=1,
)
Then we compute region signals and extract useful phenotypic information.
children = []
pooled_subjects = []
groups = [] # child or adult
for func_file, confound_file, phenotype in zip(
development_dataset.func,
development_dataset.confounds,
development_dataset.phenotypic["Child_Adult"],
strict=False,
):
time_series = masker.fit_transform(func_file, confounds=confound_file)
pooled_subjects.append(time_series)
if phenotype == "child":
children.append(time_series)
groups.append(phenotype)
print(f"Data has {len(children)} children.")
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
________________________________________________________________________________
[Memory] Calling nilearn.image.resampling.resample_img...
resample_img(<nibabel.nifti1.Nifti1Image object at 0x7f36362bd0f0>, interpolation='linear', target_shape=(50, 59, 50), target_affine=array([[ 4., 0., 0., -96.],
[ 0., 4., 0., -132.],
[ 0., 0., 4., -78.],
[ 0., 0., 0., 1.]]))
_____________________________________________________resample_img - 0.2s, 0.0min
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3611bf2410>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar128_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.8s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3611bf36d0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362bf520>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3611bf3a00>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.8s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634533fa0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.8s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3611bf3cd0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar127_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634530640>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar024_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362be4a0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar023_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3612131210>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar022_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362be6e0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar021_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362bd780>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar020_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362bd780>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar019_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3612131210>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar018_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36362bd780>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar017_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3611bf22c0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.8s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36104ff160>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36104fe350>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36345301f0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36345301f0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634532290>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36345308e0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634532290>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3612133f40>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634532290>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36104fd630>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3634532290>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3612133f40>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3619211de0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f36104fd630>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
[NiftiMapsMasker.wrapped] Loading regions from
'/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Resampling regions
[NiftiMapsMasker.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7f3619211de0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': False,
'keep_masked_maps': False,
'low_pass': 0.1,
'maps_img': '/home/runner/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': False,
'standardize_confounds': True,
't_r': 2,
'target_affine': None,
'target_shape': None}, confounds=[ '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_group_level_connectivity.py:67: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(func_file, confounds=confound_file)
_______________________________________________filter_and_extract - 0.7s, 0.0min
Data has 24 children.
ROI-to-ROI correlations of children¶
The simpler and most commonly used kind of connectivity is correlation. It
models the full (marginal) connectivity between pairwise ROIs. We can
estimate it using ConnectivityMeasure.
from nilearn.connectome import ConnectivityMeasure
correlation_measure = ConnectivityMeasure(kind="correlation", verbose=1)
From the list of ROIs time-series for children, the correlation_measure computes individual correlation matrices.
correlation_matrices = correlation_measure.fit_transform(children)
# All individual coefficients are stacked in a unique 2D matrix.
print(
"Correlations of children are stacked "
f"in an array of shape {correlation_matrices.shape}"
)
[ConnectivityMeasure.wrapped] Finished fit
Correlations of children are stacked in an array of shape (24, 39, 39)
as well as the average correlation across all fitted subjects.
mean_correlation_matrix = correlation_measure.mean_
print(f"Mean correlation has shape {mean_correlation_matrix.shape}.")
Mean correlation has shape (39, 39).
We display the connectome matrices of the first 3 children
import numpy as np
from matplotlib import pyplot as plt
_, axes = plt.subplots(1, 3, figsize=(15, 5))
vmax = np.absolute(correlation_matrices).max()
for i, (matrix, ax) in enumerate(
zip(correlation_matrices, axes, strict=False)
):
plot_matrix(
matrix,
tri="lower",
axes=ax,
title=f"correlation, child {i}",
vmax=vmax,
vmin=-vmax,
)
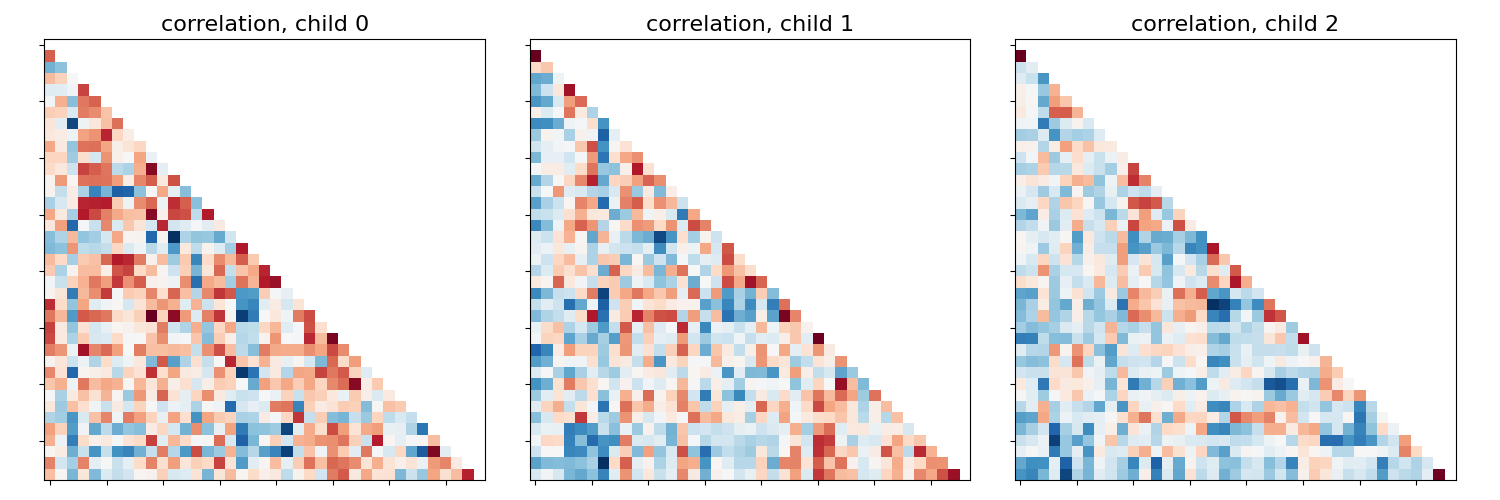
The blocks structure that reflect functional networks are visible.
Now we display as a connectome the mean correlation matrix over all children.
plot_connectome(
mean_correlation_matrix,
msdl_coords,
title="mean correlation over all children",
)
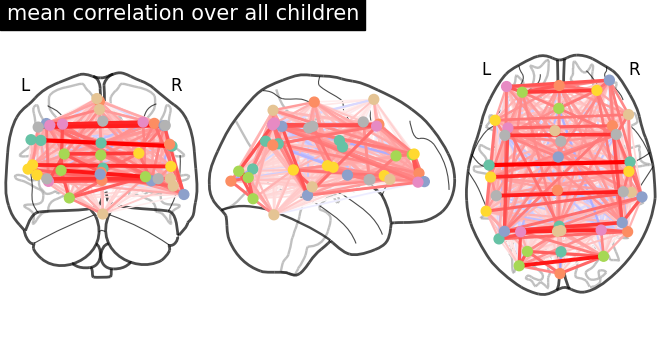
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f35cdc6e170>
Studying partial correlations¶
We can also study direct connections, revealed by partial correlation coefficients. We just change the ConnectivityMeasure kind
partial_correlation_measure = ConnectivityMeasure(
kind="partial correlation", verbose=1
)
partial_correlation_matrices = partial_correlation_measure.fit_transform(
children
)
[ConnectivityMeasure.wrapped] Finished fit
Most of direct connections are weaker than full connections.
_, axes = plt.subplots(1, 3, figsize=(15, 5))
vmax = np.absolute(partial_correlation_matrices).max()
for i, (matrix, ax) in enumerate(
zip(partial_correlation_matrices, axes, strict=False)
):
plot_matrix(
matrix,
tri="lower",
axes=ax,
title=f"partial correlation, child {i}",
vmax=vmax,
vmin=-vmax,
)
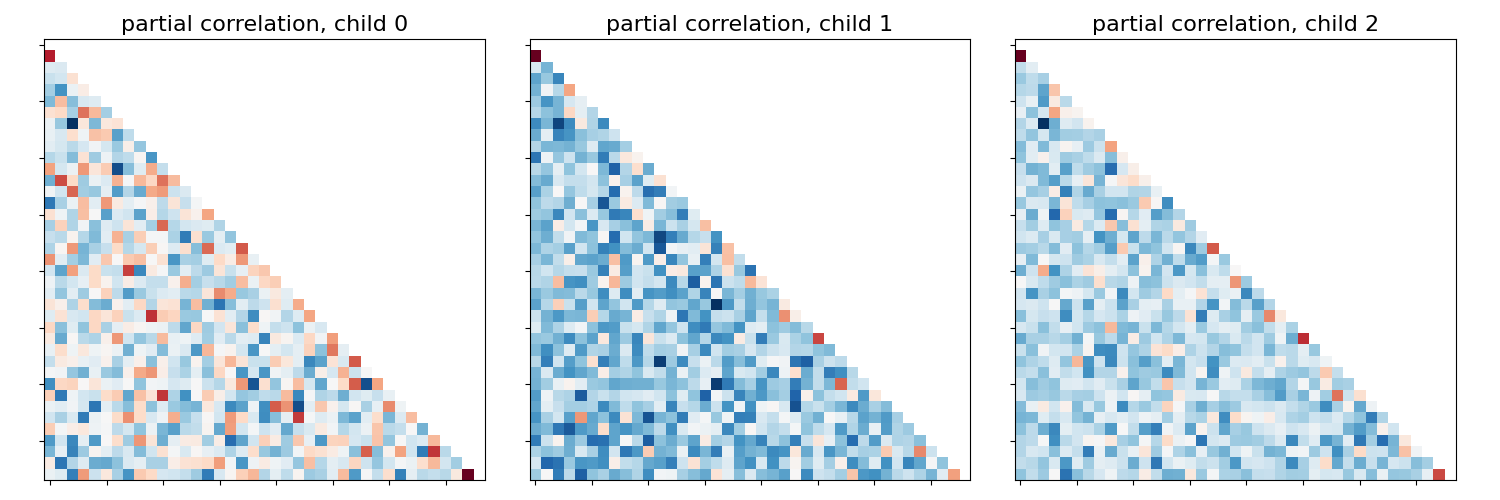
plot_connectome(
partial_correlation_measure.mean_,
msdl_coords,
title="mean partial correlation over all children",
)
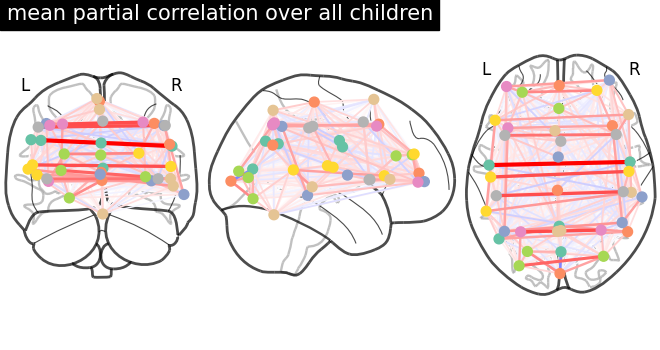
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f3610d1dc60>
Extract subjects variabilities around a group connectivity¶
We can use both correlations and partial correlations to capture reproducible connectivity patterns at the group-level. This is done by the tangent space embedding.
tangent_measure = ConnectivityMeasure(kind="tangent", verbose=1)
We fit our children group and get the group connectivity matrix stored as in tangent_measure.mean_, and individual deviation matrices of each subject from it.
tangent_matrices model individual connectivities as perturbations of the group connectivity matrix tangent_measure.mean_. Keep in mind that these subjects-to-group variability matrices do not directly reflect individual brain connections. For instance negative coefficients can not be interpreted as anticorrelated regions.
_, axes = plt.subplots(1, 3, figsize=(15, 5))
for i, (matrix, ax) in enumerate(zip(tangent_matrices, axes, strict=False)):
plot_matrix(
matrix,
tri="lower",
axes=ax,
title=f"tangent offset, child {i}",
)
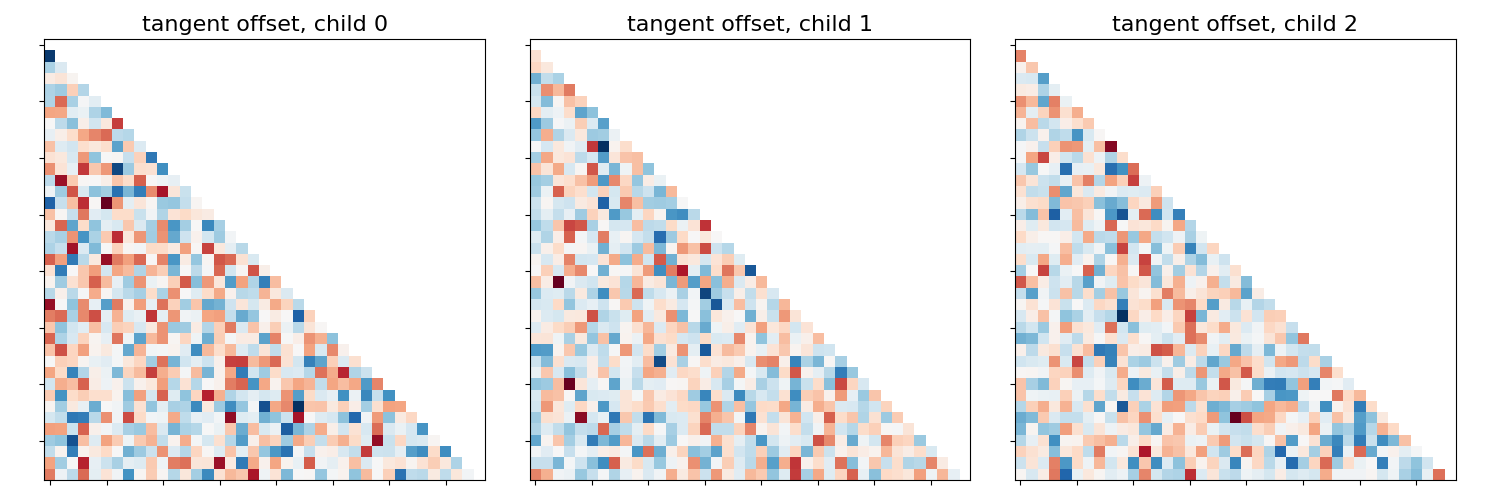
The average tangent matrix cannot be interpreted, as individual matrices represent deviations from the mean, which is set to 0.
What kind of connectivity is most powerful for classification?¶
We will use connectivity matrices as features to distinguish children from adults. We use cross-validation and measure classification accuracy to compare the different kinds of connectivity matrices. We use random splits of the subjects into training/testing sets. StratifiedShuffleSplit allows preserving the proportion of children in the test set.
from sklearn.metrics import accuracy_score
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.svm import LinearSVC
kinds = ["correlation", "partial correlation", "tangent"]
_, classes = np.unique(groups, return_inverse=True)
cv = StratifiedShuffleSplit(n_splits=15, random_state=0, test_size=5)
pooled_subjects = np.asarray(pooled_subjects)
scores = {}
for kind in kinds:
scores[kind] = []
for train, test in cv.split(pooled_subjects, classes):
# *ConnectivityMeasure* can output the estimated subjects coefficients
# as a 1D arrays through the parameter *vectorize*.
connectivity = ConnectivityMeasure(
kind=kind, vectorize=True, verbose=1
)
# build vectorized connectomes for subjects in the train set
connectomes = connectivity.fit_transform(pooled_subjects[train])
# fit the classifier
classifier = LinearSVC(dual=True).fit(connectomes, classes[train])
# make predictions for the left-out test subjects
predictions = classifier.predict(
connectivity.transform(pooled_subjects[test])
)
# store the accuracy for this cross-validation fold
scores[kind].append(accuracy_score(classes[test], predictions))
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
display the results
mean_scores = [np.mean(scores[kind]) for kind in kinds]
scores_std = [np.std(scores[kind]) for kind in kinds]
plt.figure(figsize=(6, 4), constrained_layout=True)
positions = np.arange(len(kinds)) * 0.1 + 0.1
plt.barh(positions, mean_scores, align="center", height=0.05, xerr=scores_std)
yticks = [k.replace(" ", "\n") for k in kinds]
plt.yticks(positions, yticks)
plt.gca().grid(True)
plt.gca().set_axisbelow(True)
plt.gca().axvline(0.8, color="red", linestyle="--")
plt.xlabel("Classification accuracy\n(red line = chance level)")
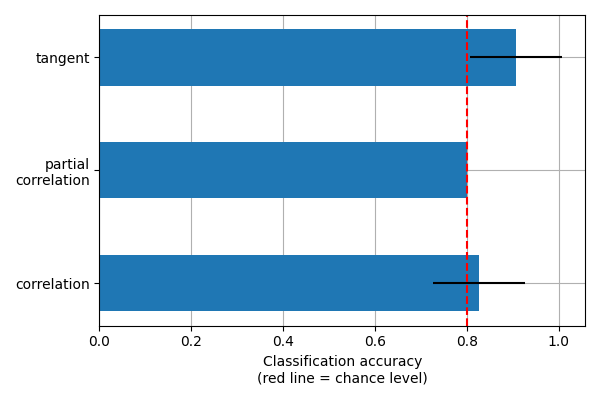
Text(0.5, 34.367, 'Classification accuracy\n(red line = chance level)')
This is a small example to showcase nilearn features. In practice such comparisons need to be performed on much larger cohorts and several datasets. Dadi et al.[1] showed that across many cohorts and clinical questions, the tangent kind should be preferred.
show()
References¶
Total running time of the script: (2 minutes 14.297 seconds)
Estimated memory usage: 952 MB