Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Controlling the contrast of the background when plotting¶
The dim argument controls the contrast of the background.
dim modifies the contrast of this image: dim=0 leaves the image unchanged, negative values of dim enhance it, and positive values decrease it (dim the background).
This dim argument may also be useful for the plot_roi function used to display ROIs on top of a background image.
Retrieve the data: the localizer dataset with contrast maps¶
from nilearn import datasets
localizer_dataset = datasets.fetch_localizer_button_task()
# Contrast map of motor task
localizer_tmap_filename = localizer_dataset.tmap
# Subject specific anatomical image
localizer_anat_filename = localizer_dataset.anat
[fetch_localizer_button_task] Dataset created in
/home/runner/nilearn_data/brainomics_localizer
[fetch_localizer_button_task] Downloading data from
https://osf.io/hwbm2/download ...
[fetch_localizer_button_task] ...done. (4 seconds, 0 min)
[fetch_localizer_button_task] Downloading data from
https://osf.io/download/5d27ddc91c5b4a001b9ef9d0/ ...
[fetch_localizer_button_task] ...done. (3 seconds, 0 min)
[fetch_localizer_button_task] Downloading data from
https://osf.io/download/5d27d29145253a001c3e284f/ ...
[fetch_localizer_button_task] ...done. (2 seconds, 0 min)
[fetch_localizer_button_task] Downloading data from
https://osf.io/download/5d275ede1c5b4a001aa00c26/ ...
[fetch_localizer_button_task] Downloaded 2236416 of 13951266 bytes (16.0%%,
5.3s remaining)
[fetch_localizer_button_task] ...done. (3 seconds, 0 min)
[fetch_localizer_button_task] Downloading data from
https://osf.io/download/5d27037f45253a001c3d4563/ ...
[fetch_localizer_button_task] ...done. (1 seconds, 0 min)
[fetch_localizer_button_task] Downloading data from
https://osf.io/download/5d7b8948fcbf44001c44e695/ ...
[fetch_localizer_button_task] ...done. (1 seconds, 0 min)
Plotting with enhancement of background image with dim=-.5¶
from nilearn.plotting import plot_stat_map, show
cut_coords = (36, -27, 66)
plot_stat_map(
localizer_tmap_filename,
bg_img=localizer_anat_filename,
cut_coords=cut_coords,
threshold=3,
title="dim=-.5",
dim=-0.5,
)
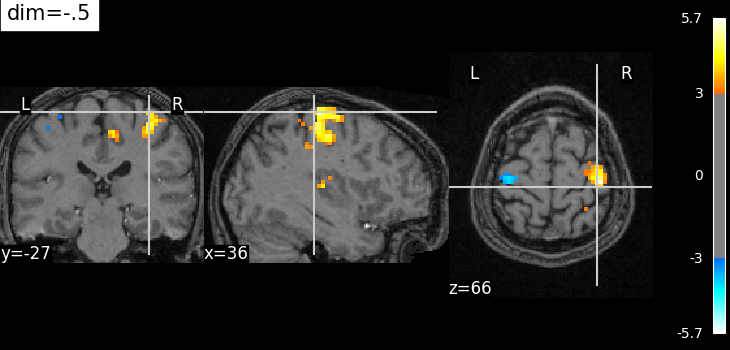
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f32c302ca90>
Plotting with no change of contrast in background image with dim=0¶
plot_stat_map(
localizer_tmap_filename,
bg_img=localizer_anat_filename,
cut_coords=cut_coords,
threshold=3,
title="dim=0",
dim=0,
)
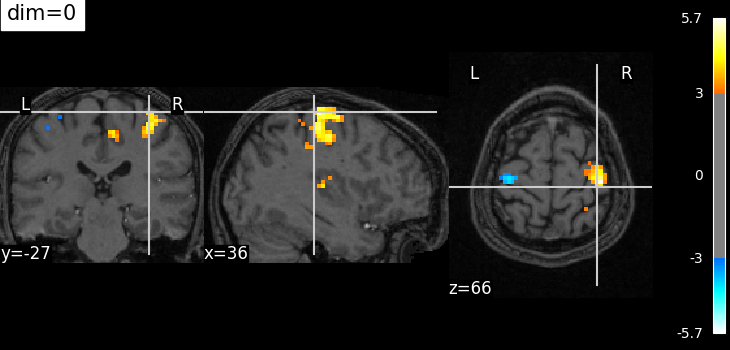
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f32c44c2530>
Plotting with decrease of contrast in background image with dim=.5¶
plot_stat_map(
localizer_tmap_filename,
bg_img=localizer_anat_filename,
cut_coords=cut_coords,
threshold=3,
title="dim=.5",
dim=0.5,
)
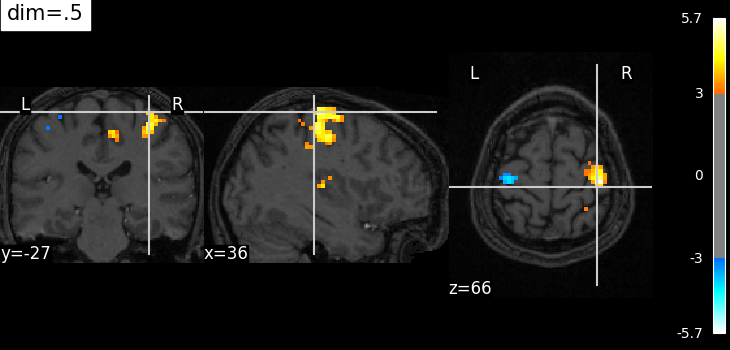
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f32c4d701f0>
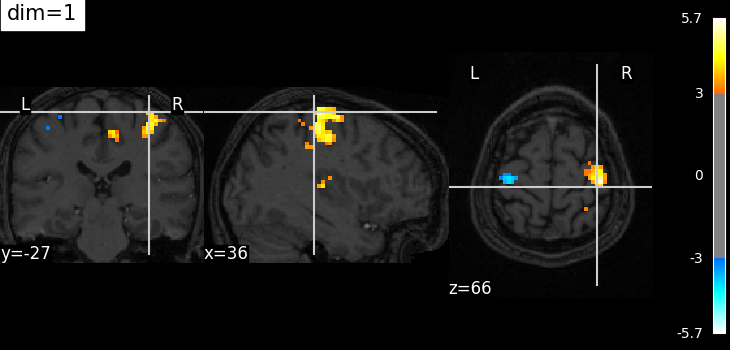
Plotting with more decrease in contrast with dim=1¶
plot_stat_map(
localizer_tmap_filename,
bg_img=localizer_anat_filename,
cut_coords=cut_coords,
threshold=3,
title="dim=1",
dim=1,
)
show()
Total running time of the script: (0 minutes 17.525 seconds)
Estimated memory usage: 108 MB