Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Comparing connectomes on different reference atlases¶
This examples shows how to turn a parcellation into connectome for
visualization. This requires choosing centers for each parcel
or network, via find_parcellation_cut_coords for
parcellation based on labels and
find_probabilistic_atlas_cut_coords for
parcellation based on probabilistic values.
In the intermediary steps, we make use of
MultiNiftiLabelsMasker and
MultiNiftiMapsMasker
to extract time series from nifti
objects from multiple subjects using different parcellation atlases.
The time series of all subjects of the brain development dataset are
concatenated and given directly to
ConnectivityMeasure for computing parcel-wise
correlation matrices for each atlas across all subjects.
Mean correlation matrix is displayed on glass brain on extracted coordinates.
Load atlases¶
from nilearn.datasets import fetch_atlas_yeo_2011, fetch_development_fmri
yeo = fetch_atlas_yeo_2011(n_networks=17)
print(
"Yeo atlas nifti image (3D) with 17 parcels and liberal mask "
f" is located at: {yeo['maps']}"
)
[fetch_atlas_yeo_2011] Dataset found in /home/runner/nilearn_data/yeo_2011
Yeo atlas nifti image (3D) with 17 parcels and liberal mask is located at: /home/runner/nilearn_data/yeo_2011/Yeo_JNeurophysiol11_MNI152/Yeo2011_17Networks_MNI152_FreeSurferConformed1mm_LiberalMask.nii.gz
Load functional data¶
data = fetch_development_fmri(n_subjects=10)
print(
"Functional nifti images (4D, e.g., one subject) "
f"are located at : {data.func[0]!r}"
)
print(
"Counfound csv files (of same subject) are located "
f"at : {data['confounds'][0]!r}"
)
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
Functional nifti images (4D, e.g., one subject) are located at : '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
Counfound csv files (of same subject) are located at : '/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_desc-reducedConfounds_regressors.tsv'
Extract coordinates on Yeo atlas - parcellations¶
from nilearn.connectome import ConnectivityMeasure
from nilearn.maskers import MultiNiftiLabelsMasker
# ConnectivityMeasure from Nilearn uses simple 'correlation' to compute
# connectivity matrices for all subjects in a list
connectome_measure = ConnectivityMeasure(kind="correlation", verbose=1)
# create masker using MultiNiftiLabelsMasker to extract functional data within
# atlas parcels from multiple subjects using parallelization to speed up the
# computation
masker = MultiNiftiLabelsMasker(
labels_img=yeo["maps"], # Both hemispheres
standardize_confounds=True,
memory="nilearn_cache",
n_jobs=2,
verbose=1,
)
# extract time series from all subjects
time_series = masker.fit_transform(data.func, confounds=data.confounds)
# calculate correlation matrices across subjects and display
correlation_matrices = connectome_measure.fit_transform(time_series)
# Mean correlation matrix across 10 subjects can be grabbed like this,
# using connectome measure object
mean_correlation_matrix = connectome_measure.mean_
# useful for plotting connectivity interactions on glass brain
from nilearn.plotting import (
find_parcellation_cut_coords,
plot_connectome,
show,
)
# grab center coordinates for atlas labels
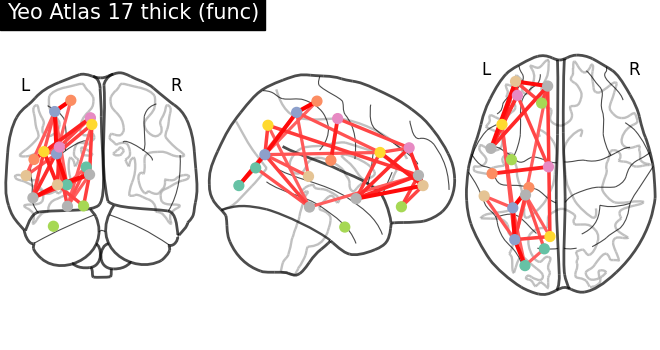
coordinates = find_parcellation_cut_coords(labels_img=yeo["maps"])
# plot connectome with 80% edge strength in the connectivity
left_connectome = plot_connectome(
mean_correlation_matrix, coordinates, edge_threshold="80%"
)
show()
[MultiNiftiLabelsMasker.fit_transform] Loading regions from
'/home/runner/nilearn_data/yeo_2011/Yeo_JNeurophysiol11_MNI152/Yeo2011_17Network
s_MNI152_FreeSurferConformed1mm_LiberalMask.nii.gz'
[MultiNiftiLabelsMasker.fit_transform] Resampling regions
[MultiNiftiLabelsMasker.fit_transform] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
Note
The approach above will extract time series and compute a single connectivity matrix for both hemispheres. However, the connectome is plotted only for the left hemisphere.
If your aim is to compute and plot hemisphere-wise connectivity, you can follow the example below.
First, create a separate atlas image for each hemisphere:
import nibabel as nb
import numpy as np
from nilearn.image import get_data, new_img_like
from nilearn.image.resampling import coord_transform
# load the atlas image first
label_image = nb.load(yeo["maps"])
# extract the affine matrix of the image
labels_affine = label_image.affine
# generate image coordinates using affine
x, y, z = coord_transform(0, 0, 0, np.linalg.inv(labels_affine))
# generate an separate image for the left hemisphere
# left/right split is done along x-axis
left_hemi = get_data(label_image).copy()
left_hemi[: int(x)] = 0
label_image_left = new_img_like(label_image, left_hemi, labels_affine)
# same for the right hemisphere
right_hemi = get_data(label_image).copy()
right_hemi[int(x) :] = 0
label_image_right = new_img_like(label_image, right_hemi, labels_affine)
Then, create a masker object, compute a connectivity matrix and plot the results for each hemisphere:
for hemi, img in zip(
["right", "left"], [label_image_right, label_image_left], strict=False
):
masker = MultiNiftiLabelsMasker(
labels_img=img,
standardize_confounds=True,
verbose=1,
)
time_series = masker.fit_transform(data.func, confounds=data.confounds)
correlation_matrices = connectome_measure.fit_transform(time_series)
mean_correlation_matrix = connectome_measure.mean_
coordinates = find_parcellation_cut_coords(
labels_img=img, label_hemisphere=hemi
)
plot_connectome(
mean_correlation_matrix,
coordinates,
edge_threshold="80%",
title=f"Yeo Atlas 17 thick (func) - {hemi}",
)
show()
[MultiNiftiLabelsMasker.fit_transform] Loading regions from
<nibabel.nifti1.Nifti1Image object at 0x7f35f9298820>
[MultiNiftiLabelsMasker.fit_transform] Resampling regions
[MultiNiftiLabelsMasker.fit_transform] Finished fit
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f35f9299ba0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f36345324d0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3636309ae0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f36345318d0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f35cdc6e860>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3634532ad0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f35ebc2a8f0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3634530fd0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f363630af80>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3634532830>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[ConnectivityMeasure.wrapped] Finished fit
[MultiNiftiLabelsMasker.fit_transform] Loading regions from
<nibabel.nifti1.Nifti1Image object at 0x7f35b2a53100>
[MultiNiftiLabelsMasker.fit_transform] Resampling regions
[MultiNiftiLabelsMasker.fit_transform] Finished fit
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f36196766e0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619600430>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619600ee0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619601660>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619602170>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619600a30>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f36196027d0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619601cf0>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619603730>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[MultiNiftiLabelsMasker.fit_transform] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f3619600f40>
[MultiNiftiLabelsMasker.fit_transform] Extracting region signals
[MultiNiftiLabelsMasker.fit_transform] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:151: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
time_series = masker.fit_transform(data.func, confounds=data.confounds)
[ConnectivityMeasure.wrapped] Finished fit
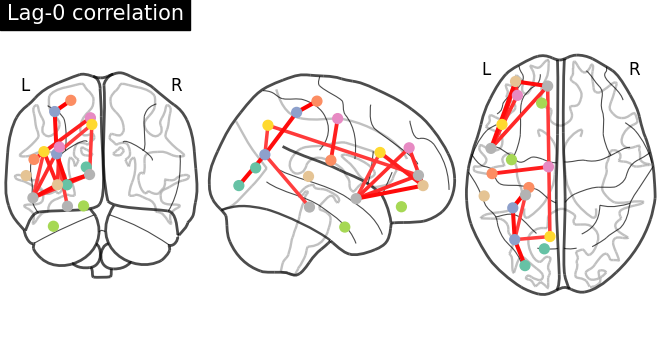
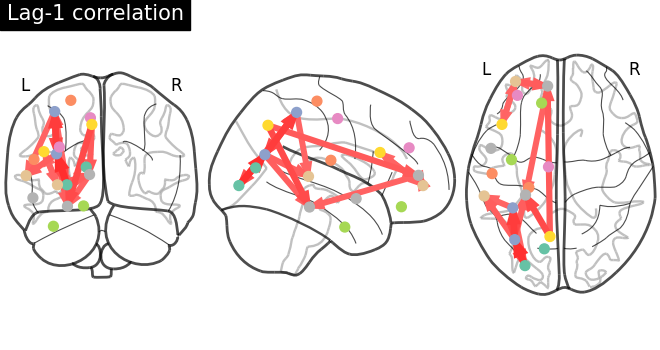
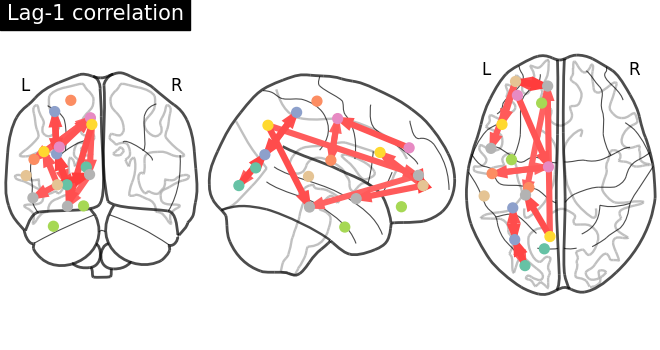
Plot a directed connectome - asymmetric connectivity measure¶
In this section, we use the lag-1 correlation as the connectivity measure, which leads to an asymmetric connectivity matrix. The plot_connectome function accepts both symmetric and asymmetric matrices, but plots the latter as a directed graph.
# Define a custom function to compute lag correlation on the time series
def lag_correlation(time_series, lag):
n_subjects = len(time_series)
_, n_features = time_series[0].shape
lag_cor = np.zeros((n_subjects, n_features, n_features))
for subject, serie in enumerate(time_series):
for i in range(n_features):
for j in range(n_features):
if lag == 0:
lag_cor[subject, i, j] = np.corrcoef(
serie[:, i], serie[:, j]
)[0, 1]
else:
lag_cor[subject, i, j] = np.corrcoef(
serie[lag:, i], serie[:-lag, j]
)[0, 1]
return np.mean(lag_cor, axis=0)
# Compute lag-0 and lag-1 correlations and plot associated connectomes
for lag in [0, 1]:
lag_correlation_matrix = lag_correlation(time_series, lag)
plot_connectome(
lag_correlation_matrix,
coordinates,
edge_threshold="90%",
title=f"Lag-{lag} correlation",
)
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_atlas_comparison.py:200: UserWarning: 'adjacency_matrix' is not symmetric.
A directed graph will be plotted.
plot_connectome(
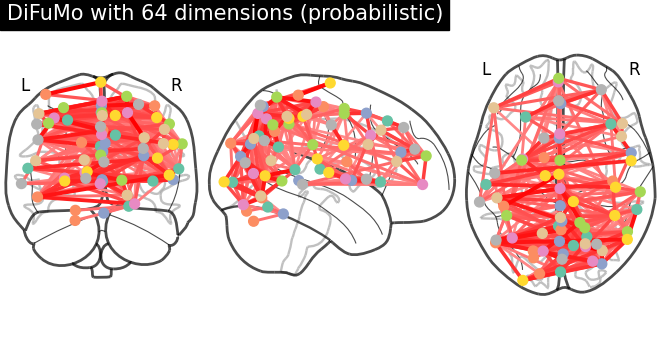
Load probabilistic atlases - extracting coordinates on brain maps¶
from nilearn.datasets import fetch_atlas_difumo
from nilearn.plotting import find_probabilistic_atlas_cut_coords
dim = 64
difumo = fetch_atlas_difumo(dimension=dim, resolution_mm=2)
[fetch_atlas_difumo] Dataset found in /home/runner/nilearn_data/difumo_atlases
Iterate over fetched atlases to extract coordinates - probabilistic¶
from nilearn.maskers import MultiNiftiMapsMasker
# Create masker using MultiNiftiMapsMasker to extract functional data within
# atlas parcels from multiple subjects using parallelization to speed up the
# computation.
masker = MultiNiftiMapsMasker(
maps_img=difumo.maps,
standardize_confounds=True,
memory="nilearn_cache",
memory_level=1,
n_jobs=2,
verbose=1,
)
# extract time series from all subjects
time_series = masker.fit_transform(data.func, confounds=data.confounds)
# calculate correlation matrices across subjects and display
correlation_matrices = connectome_measure.fit_transform(time_series)
# Mean correlation matrix across 10 subjects can be grabbed like this,
# using connectome measure object
mean_correlation_matrix = connectome_measure.mean_
# grab center coordinates for probabilistic atlas
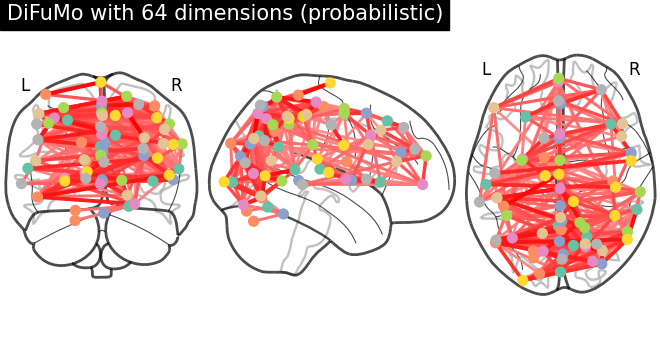
coordinates = find_probabilistic_atlas_cut_coords(maps_img=difumo.maps)
# plot connectome with 85% edge strength in the connectivity
plot_connectome(
mean_correlation_matrix,
coordinates,
edge_threshold="85%",
title=f"DiFuMo with {dim} dimensions (probabilistic)",
)
show()
[MultiNiftiMapsMasker.fit_transform] Loading regions from
'/home/runner/nilearn_data/difumo_atlases/64/2mm/maps.nii.gz'
[MultiNiftiMapsMasker.fit_transform] Resampling regions
________________________________________________________________________________
[Memory] Calling nilearn.image.resampling.resample_img...
resample_img(<nibabel.nifti1.Nifti1Image object at 0x7f3634265a20>, interpolation='linear', target_shape=(50, 59, 50), target_affine=array([[ 4., 0., 0., -96.],
[ 0., 4., 0., -132.],
[ 0., 0., 4., -78.],
[ 0., 0., 0., 1.]]))
_____________________________________________________resample_img - 0.5s, 0.0min
[MultiNiftiMapsMasker.fit_transform] Finished fit
[ConnectivityMeasure.wrapped] Finished fit
Total running time of the script: (1 minutes 31.866 seconds)
Estimated memory usage: 2831 MB