Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Regions extraction using dictionary learning and functional connectomes¶
This example shows how to use RegionExtractor
to extract spatially constrained brain regions from whole brain maps decomposed
using Dictionary learning and use them to build
a functional connectome.
We used 20 movie-watching functional datasets from
fetch_development_fmri and
DictLearning for set of brain atlas maps.
This example can also be inspired to apply the same steps
to even regions extraction
using ICA maps.
In that case, idea would be to replace
Dictionary learning to canonical ICA decomposition
using CanICA
Please see the related documentation
of RegionExtractor for more details.
Fetch brain development functional datasets¶
We use nilearn’s datasets downloading utilities
from nilearn.datasets import fetch_development_fmri
rest_dataset = fetch_development_fmri(n_subjects=20)
func_filenames = rest_dataset.func
confounds = rest_dataset.confounds
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in
/home/runner/nilearn_data/development_fmri/development_fmri
Extract functional networks with Dictionary learning¶
Import DictLearning from the
decomposition module, instantiate the object, and
fit the model to the
functional datasets
from nilearn.decomposition import DictLearning
# Initialize DictLearning object
dict_learn = DictLearning(
n_components=8,
smoothing_fwhm=6.0,
memory="nilearn_cache",
memory_level=1,
random_state=0,
verbose=1,
)
# Fit to the data
dict_learn.fit(func_filenames)
# Resting state networks/maps in attribute `components_img_`
components_img = dict_learn.components_img_
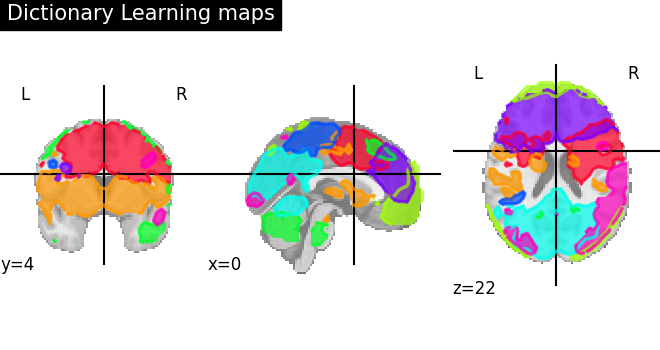
# Visualization of functional networks
# Show networks using plotting utilities
from nilearn.plotting import plot_prob_atlas, show
plot_prob_atlas(
components_img,
view_type="filled_contours",
title="Dictionary Learning maps",
draw_cross=False,
)
show()
[DictLearning.fit] Loading data from
['/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-
pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz']
[DictLearning.fit] Computing mask
[DictLearning.fit] Resampling mask
[DictLearning.fit] Finished fit
[DictLearning.fit] Loading data
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f35f991b4f0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bee530>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610becac0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bef460>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610becf10>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bee4d0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bef190>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610beff40>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610befca0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bef7f0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bee4d0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bed5a0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bec820>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610befcd0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f35f94ba0e0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f35f94ba9b0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f35f94ba650>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bec970>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610befbb0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f3610bed7b0>
[DictLearning.fit] Smoothing images
[DictLearning.fit] Extracting region signals
[DictLearning.fit] Cleaning extracted signals
[DictLearning.fit] Learning initial components
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 1.3s finished
[DictLearning.fit] Computing initial loadings
________________________________________________________________________________
[Memory] Calling nilearn.decomposition.dict_learning._compute_loadings...
_compute_loadings(array([[0.00061 , ..., 0.002672],
...,
[0.005603, ..., 0.000519]], shape=(8, 28267)),
array([[-3.895898, ..., -0.034474],
...,
[ 1.466817, ..., -0.956513]], shape=(160, 28267)))
_________________________________________________compute_loadings - 0.0s, 0.0min
[DictLearning.fit] Learning dictionary
________________________________________________________________________________
[Memory] Calling sklearn.decomposition._dict_learning.dict_learning_online...
dict_learning_online(array([[-3.895898, ..., 1.466817],
...,
[-0.034474, ..., -0.956513]], shape=(28267, 160)),
8, alpha=10, batch_size=20, method='cd', dict_init=array([[ 0.091985, ..., 0.142599],
...,
[-0.202624, ..., -0.229184]], shape=(8, 160)), verbose=0, random_state=0, return_code=True, shuffle=True, n_jobs=1, max_iter=1414)
_____________________________________________dict_learning_online - 1.2s, 0.0min
[DictLearning.fit] Computing image from signals
Extract regions from networks¶
Import RegionExtractor from the
regions module.
threshold=0.5 indicates that we keep nominal of amount nonzero
voxels across all maps, less the threshold means that
more intense non-voxels will be survived.
from nilearn.regions import RegionExtractor
extractor = RegionExtractor(
components_img,
threshold=0.5,
thresholding_strategy="ratio_n_voxels",
extractor="local_regions",
standardize_confounds=True,
min_region_size=1350,
verbose=1,
)
# Just call fit() to process for regions extraction
extractor.fit()
# Extracted regions are stored in regions_img_
regions_extracted_img = extractor.regions_img_
# Each region index is stored in index_
regions_index = extractor.index_
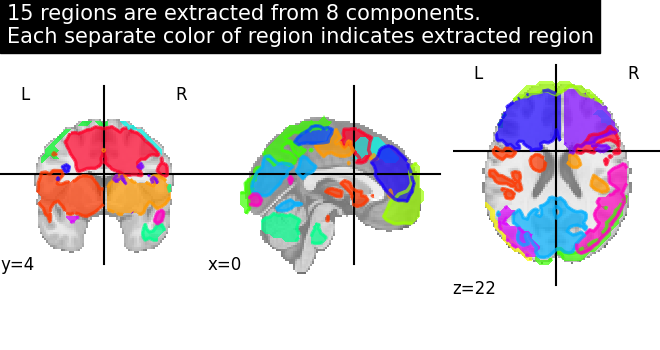
# Total number of regions extracted
n_regions_extracted = regions_extracted_img.shape[-1]
# Visualization of region extraction results
title = (
f"{n_regions_extracted} regions are extracted from 8 components.\n"
"Each separate color of region indicates extracted region"
)
plot_prob_atlas(
regions_extracted_img,
view_type="filled_contours",
title=title,
draw_cross=False,
)
show()
[RegionExtractor.fit] Loading regions from <nibabel.nifti1.Nifti1Image object at
0x7f35cf012b60>
[RegionExtractor.fit] Finished fit
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/ma/core.py:2892: UserWarning: Warning: converting a masked element to nan.
_data = np.array(data, dtype=dtype, copy=copy,
Compute correlation coefficients¶
First we need to do subjects timeseries signals extraction
and then estimating correlation matrices on those signals.
To extract timeseries signals, we call
transform onto each subject
functional data stored in func_filenames.
To estimate correlation matrices we import connectome utilities from nilearn.
from nilearn.connectome import ConnectivityMeasure
correlations = []
# Initializing ConnectivityMeasure object with kind='correlation'
connectome_measure = ConnectivityMeasure(kind="correlation", verbose=1)
for filename, confound in zip(func_filenames, confounds, strict=False):
# call transform from RegionExtractor object to extract timeseries signals
timeseries_each_subject = extractor.transform(filename, confounds=confound)
# call fit_transform from ConnectivityMeasure object
correlation = connectome_measure.fit_transform([timeseries_each_subject])
# saving each subject correlation to correlations
correlations.append(correlation)
# Mean of all correlations
import numpy as np
mean_correlations = np.mean(correlations, axis=0).reshape(
n_regions_extracted, n_regions_extracted
)
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar126_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar124_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar125_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar016_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar015_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar014_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar013_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar012_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar011_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar008_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar007_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar006_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar005_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar004_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar009_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[RegionExtractor.wrapped] Loading data from
'/home/runner/nilearn_data/development_fmri/development_fmri/sub-pixar010_task-p
ixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[RegionExtractor.wrapped] Smoothing images
[RegionExtractor.wrapped] Extracting region signals
[RegionExtractor.wrapped] Cleaning extracted signals
/home/runner/work/nilearn/nilearn/examples/03_connectivity/plot_extract_regions_dictlearning_maps.py:134: FutureWarning: boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
timeseries_each_subject = extractor.transform(filename, confounds=confound)
[ConnectivityMeasure.wrapped] Finished fit
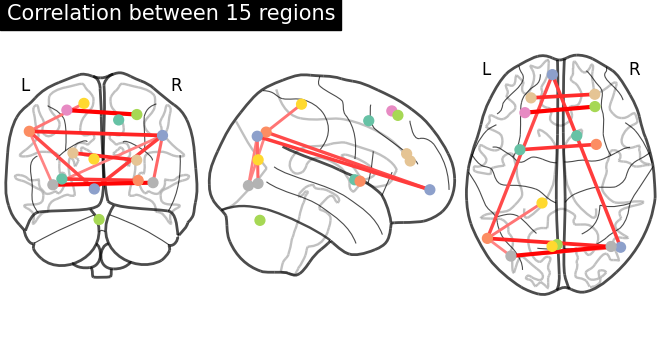
Plot resulting connectomes¶
First we plot the mean of correlation matrices with
plot_matrix, and we use
plot_connectome to plot the
connectome relations.
from nilearn.plotting import (
find_probabilistic_atlas_cut_coords,
find_xyz_cut_coords,
plot_connectome,
plot_matrix,
)
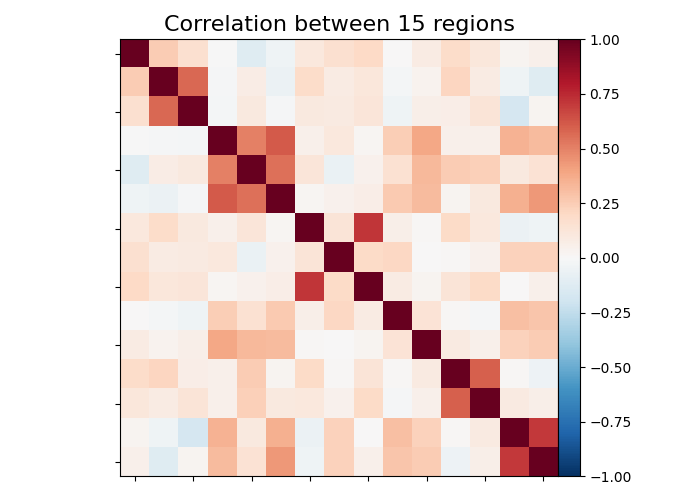
title = f"Correlation between {int(n_regions_extracted)} regions"
# First plot the matrix
plot_matrix(mean_correlations, vmax=1, vmin=-1, title=title)
# Then find the center of the regions and plot a connectome
regions_img = regions_extracted_img
coords_connectome = find_probabilistic_atlas_cut_coords(regions_img)
plot_connectome(
mean_correlations, coords_connectome, edge_threshold="90%", title=title
)
show()
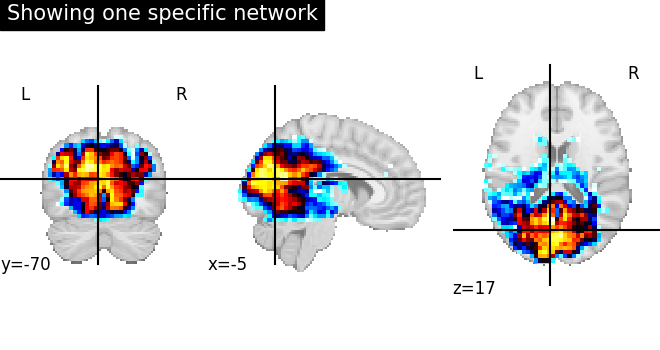
Plot regions extracted for only one specific network¶
First, we plot a network of index=4
without region extraction (left plot).
from nilearn import image
from nilearn.plotting import plot_stat_map
img = image.index_img(components_img, 4)
coords = find_xyz_cut_coords(img)
plot_stat_map(
img,
cut_coords=coords,
title="Showing one specific network",
)
show()
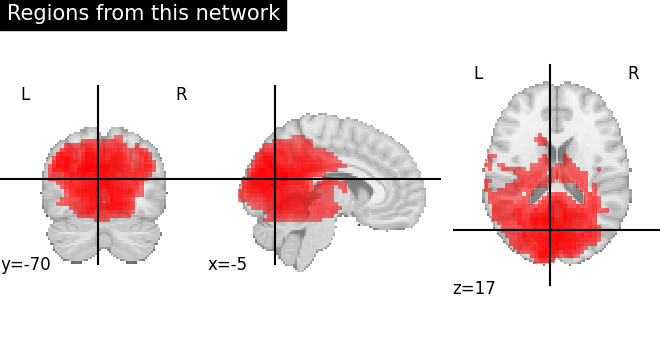
Now, we plot (right side) same network after region extraction to show that connected regions are nicely separated. Each brain extracted region is identified as separate color.
For this, we take the indices of the all regions extracted related to original network given as 4.
from nilearn.plotting import cm, plot_anat
regions_indices_of_map3 = np.where(np.array(regions_index) == 4)
display = plot_anat(
cut_coords=coords, title="Regions from this network", colorbar=False
)
# Add as an overlay all the regions of index 4
colors = "rgbcmyk"
for each_index_of_map3, color in zip(
regions_indices_of_map3[0], colors, strict=False
):
display.add_overlay(
image.index_img(regions_extracted_img, each_index_of_map3),
cmap=cm.alpha_cmap(color),
)
show()
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/ma/core.py:2892: UserWarning: Warning: converting a masked element to nan.
_data = np.array(data, dtype=dtype, copy=copy,
Total running time of the script: (0 minutes 57.251 seconds)
Estimated memory usage: 4359 MB