Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Simple example of two-runs fMRI model fitting¶
Here, we will go through a full step-by-step example of fitting a GLM to experimental data and visualizing the results. This is done on two runs of one subject of the FIAC dataset.
Here are the steps we will go through:
Set up the GLM
Compare run-specific and fixed effects contrasts
Compute a range of contrasts across both runs
Generate a report
Technically, this example shows how to handle two runs that contain the same experimental conditions. The model directly returns a fixed effect of the statistics across the two runs.
See also
See the dataset description for more information on the data used in this example.
Create an output results in the current working directory.
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_two_runs_model"
output_dir.mkdir(exist_ok=True, parents=True)
print(f"Output will be saved to: {output_dir}")
Output will be saved to: /home/runner/work/nilearn/nilearn/examples/04_glm_first_level/results/plot_two_runs_model
Set up the GLM¶
Inspecting ‘data’, we note that there are two runs. We will retain those two runs in a list of 4D img objects.
from nilearn.datasets.func import fetch_fiac_first_level
data = fetch_fiac_first_level()
fmri_imgs = [data["func1"], data["func2"]]
[fetch_fiac_first_level] Dataset found in
/home/runner/nilearn_data/fiac_nilearn.glm
Create a mean image for plotting purpose.
The design matrices were pre-computed, we simply put them in a list of DataFrames.
import numpy as np
design_matrices = [data["design_matrix1"], data["design_matrix2"]]
Initialize and run the GLM¶
First, we need to specify the model before fitting it to the data. Note that a brain mask was provided in the dataset, so that is what we will use.
from nilearn.glm.first_level import FirstLevelModel
fmri_glm = FirstLevelModel(
mask_img=data["mask"], smoothing_fwhm=5, minimize_memory=True, verbose=1
)
Compare run-specific and fixed effects contrasts¶
We can then compare run-specific and fixed effects. Here, we compare the activation produced from each run separately and then the fixed effects version.
contrast_id = "DSt_minus_SSt"
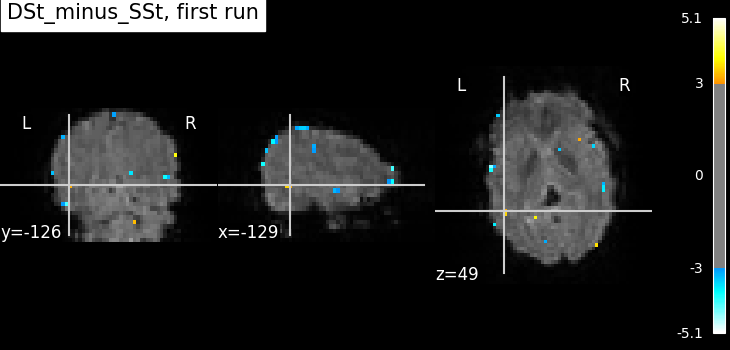
Compute the statistics for the first run.
Here, we define the contrast of interest for the first run. This may differ across runs depending on if the design matrices vary.
from nilearn.plotting import plot_stat_map, show
contrast_val = [[-1, -1, 1, 1]]
fmri_glm_run_1 = fmri_glm.fit(fmri_imgs[0], design_matrices=design_matrices[0])
summary_statistics_run_1 = fmri_glm_run_1.compute_contrast(
contrast_val,
output_type="all",
)
# Let's use the same plotting range and slices for all plots.
threshold = 3
vmax = 6.0
cut_coords = [-129, -126, 49]
plot_stat_map(
summary_statistics_run_1["z_score"],
bg_img=mean_img_,
threshold=threshold,
cut_coords=cut_coords,
title=f"{contrast_id}, first run",
vmax=vmax,
)
show()
[FirstLevelModel.fit] Loading mask from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/mask.n
ii.gz'
[FirstLevelModel.fit] Loading data from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/run1.n
ii.gz'
[FirstLevelModel.fit] Resampling mask
[FirstLevelModel.fit] Finished fit
[FirstLevelModel.fit] Computing run 1 out of 1 runs (go take a coffee, a big
one).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f326c410220>
[FirstLevelModel.fit] Smoothing images
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 1 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 0 seconds.
[FirstLevelModel.fit] Computation of 1 runs done in 1 seconds.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:91: UserWarning:
t contrasts should be of length P=13, but it has length 4. The rest of the contrast was padded with zeros.
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
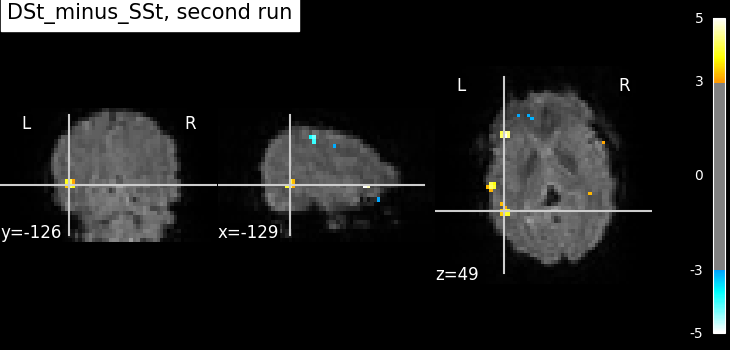
Compute the statistics for the second run.
fmri_glm_run_2 = fmri_glm.fit(fmri_imgs[1], design_matrices=design_matrices[1])
contrast_val = np.array([[-1, -1, 1, 1]])
summary_statistics_run_2 = fmri_glm_run_2.compute_contrast(
contrast_val,
output_type="all",
)
plot_stat_map(
summary_statistics_run_2["z_score"],
bg_img=mean_img_,
threshold=threshold,
cut_coords=cut_coords,
title=f"{contrast_id}, second run",
vmax=vmax,
)
show()
[FirstLevelModel.fit] Loading mask from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/mask.n
ii.gz'
[FirstLevelModel.fit] Loading data from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/run2.n
ii.gz'
[FirstLevelModel.fit] Resampling mask
[FirstLevelModel.fit] Finished fit
[FirstLevelModel.fit] Computing run 1 out of 1 runs (go take a coffee, a big
one).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f326c410190>
[FirstLevelModel.fit] Smoothing images
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 1 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 0 seconds.
[FirstLevelModel.fit] Computation of 1 runs done in 1 seconds.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:118: UserWarning:
t contrasts should be of length P=13, but it has length 4. The rest of the contrast was padded with zeros.
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
[FirstLevelModel.compute_contrast] Computing image from signals
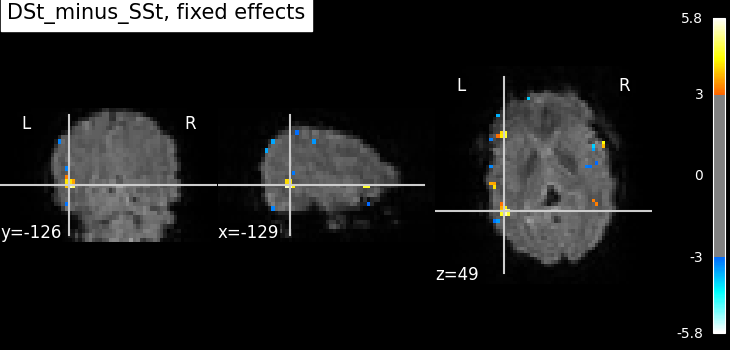
Compute the fixed effects statistics using the statistical maps of both runs.
We can use compute_fixed_effects to compute
the fixed effects statistics using the outputs
from the run-specific FirstLevelModel results.
from nilearn.glm.contrasts import compute_fixed_effects
contrast_imgs = [
summary_statistics_run_1["effect_size"],
summary_statistics_run_2["effect_size"],
]
variance_imgs = [
summary_statistics_run_1["effect_variance"],
summary_statistics_run_2["effect_variance"],
]
fixed_fx_contrast, fixed_fx_variance, fixed_fx_stat, _ = compute_fixed_effects(
contrast_imgs, variance_imgs, data["mask"]
)
plot_stat_map(
fixed_fx_stat,
bg_img=mean_img_,
threshold=threshold,
cut_coords=cut_coords,
title=f"{contrast_id}, fixed effects",
vmax=vmax,
)
show()
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:151: FutureWarning:
boolean values for 'standardize' will be deprecated in nilearn 0.15.0.
Use 'zscore_sample' instead of 'True' or use 'None' instead of 'False'.
Not unexpectedly, the fixed effects version displays higher peaks than the input runs. Computing fixed effects enhances the signal-to-noise ratio of the resulting brain maps.
Compute fixed effects statistics using preprocessed data of both runs¶
A more straightforward alternative to fitting run-specific GLMs,
than combining the results with compute_fixed_effects,
is to simply fit the GLM to both runs at once.
Since we can assume that the design matrices of both runs have the same columns, in the same order, we can again reuse the first run’s contrast vector.
[FirstLevelModel.fit] Loading mask from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/mask.n
ii.gz'
[FirstLevelModel.fit] Loading data from
'/home/runner/nilearn_data/fiac_nilearn.glm/nipy-data-0.2/data/fiac/fiac0/run1.n
ii.gz'
[FirstLevelModel.fit] Resampling mask
[FirstLevelModel.fit] Finished fit
[FirstLevelModel.fit] Computing run 1 out of 2 runs (go take a coffee, a big
one).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f32878cefb0>
[FirstLevelModel.fit] Smoothing images
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 1 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 0 seconds.
[FirstLevelModel.fit] Computing run 2 out of 2 runs (1 seconds remaining).
[FirstLevelModel.fit] Performing mask computation.
[FirstLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f326c4102e0>
[FirstLevelModel.fit] Smoothing images
[FirstLevelModel.fit] Extracting region signals
[FirstLevelModel.fit] Cleaning extracted signals
[FirstLevelModel.fit] Masking took 1 seconds.
[FirstLevelModel.fit] Performing GLM computation.
[FirstLevelModel.fit] GLM took 0 seconds.
[FirstLevelModel.fit] Computation of 2 runs done in 3 seconds.
We can just define the contrast array for one run and assume that the design matrix is the same for the other. However, if we want to be safe, we should define each contrast separately, and provide it as a list.
contrast_val = [
np.array([[-1, -1, 1, 1]]), # run 1
np.array([[-1, -1, 1, 1]]), # run 2
]
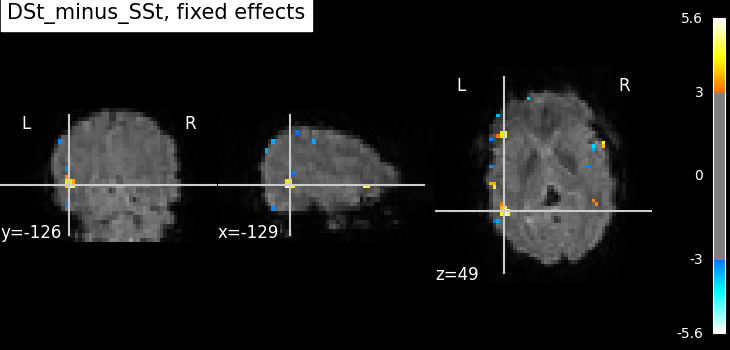
z_map = fmri_glm_multirun.compute_contrast(
contrast_val,
output_type="z_score",
)
plot_stat_map(
z_map,
bg_img=mean_img_,
threshold=threshold,
cut_coords=cut_coords,
title=f"{contrast_id}, fixed effects",
vmax=vmax,
)
show()
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:195: UserWarning:
t contrasts should be of length P=13, but it has length 4. The rest of the contrast was padded with zeros.
[FirstLevelModel.compute_contrast] Computing image from signals
You may note that the results are the same as the first fixed effects analysis, but with a lot less code.
Compute a range of contrasts across both runs¶
It may be useful to investigate a number of contrasts. Therefore, we will move beyond the original contrast of interest and both define and compute several.
Contrast specification
n_columns = design_matrices[0].shape[1]
contrasts = {
"SStSSp_minus_DStDSp": np.array([[1, 0, 0, -1]]),
"DStDSp_minus_SStSSp": np.array([[-1, 0, 0, 1]]),
"DSt_minus_SSt": np.array([[-1, -1, 1, 1]]),
"DSp_minus_SSp": np.array([[-1, 1, -1, 1]]),
"DSt_minus_SSt_for_DSp": np.array([[0, -1, 0, 1]]),
"DSp_minus_SSp_for_DSt": np.array([[0, 0, -1, 1]]),
"Deactivation": np.array([[-1, -1, -1, -1, 4]]),
"Effects_of_interest": np.eye(n_columns)[:5, :], # An F-contrast
}
Next, we compute and plot the statistics for these new contrasts.
print("Computing contrasts...")
for index, (contrast_id, contrast_val) in enumerate(contrasts.items()):
print(f" Contrast {index + 1:02g} out of {len(contrasts)}: {contrast_id}")
# Estimate the contasts.
z_map = fmri_glm.compute_contrast(contrast_val, output_type="z_score")
# Write the resulting stat images to file.
z_image_path = output_dir / f"{contrast_id}_z_map.nii.gz"
z_map.to_filename(z_image_path)
Computing contrasts...
Contrast 01 out of 8: SStSSp_minus_DStDSp
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:242: RuntimeWarning:
The same contrast will be used for all 2 runs. If the design matrices are not the same for all runs, (for example with different column names or column order across runs) you should pass contrast as an expression using the name of the conditions as they appear in the design matrices.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:242: UserWarning:
t contrasts should be of length P=13, but it has length 4. The rest of the contrast was padded with zeros.
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 02 out of 8: DStDSp_minus_SStSSp
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 03 out of 8: DSt_minus_SSt
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 04 out of 8: DSp_minus_SSp
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 05 out of 8: DSt_minus_SSt_for_DSp
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 06 out of 8: DSp_minus_SSp_for_DSt
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 07 out of 8: Deactivation
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:242: UserWarning:
t contrasts should be of length P=13, but it has length 5. The rest of the contrast was padded with zeros.
[FirstLevelModel.compute_contrast] Computing image from signals
Contrast 08 out of 8: Effects_of_interest
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:242: UserWarning:
Running approximate fixed effects on F statistics.
[FirstLevelModel.compute_contrast] Computing image from signals
Generating a report¶
Since we have already computed the FirstLevelModel and have a number of contrasts, we can quickly create a summary report.
report = fmri_glm_multirun.generate_report(
contrasts,
bg_img=mean_img_,
title="two-runs fMRI model fitting",
)
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: RuntimeWarning:
The same contrast will be used for all 2 runs. If the design matrices are not the same for all runs, (for example with different column names or column order across runs) you should pass contrast as an expression using the name of the conditions as they appear in the design matrices.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
t contrasts should be of length P=13, but it has length 4. The rest of the contrast was padded with zeros.
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Computing image from signals
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
t contrasts should be of length P=13, but it has length 5. The rest of the contrast was padded with zeros.
[FirstLevelModel.generate_report] Computing image from signals
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Running approximate fixed effects on F statistics.
[FirstLevelModel.generate_report] Computing image from signals
[FirstLevelModel.generate_report] Generating contrast-level figures...
[FirstLevelModel.generate_report] Generating design matrices figures...
[FirstLevelModel.generate_report] Generating contrast matrices figures...
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 8 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 9 column(s) of zeros.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_two_runs_model.py:254: UserWarning:
Contrasts will be padded with 8 column(s) of zeros.
Note
The generated report can be:
displayed in a Notebook,
opened in a browser using the
.open_in_browser()method,or saved to a file using the
.save_as_html(output_filepath)method.
Total running time of the script: (0 minutes 25.216 seconds)
Estimated memory usage: 629 MB