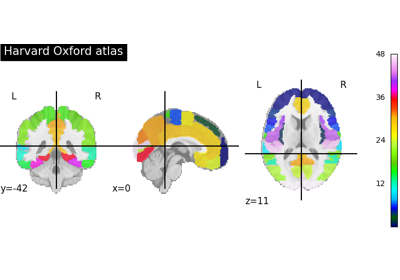
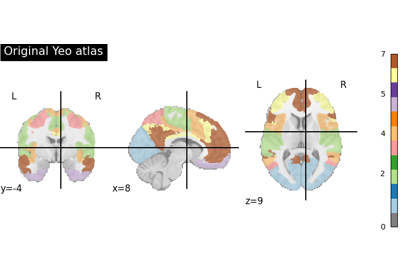
Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_yeo_2011¶
- nilearn.datasets.fetch_atlas_yeo_2011(data_dir=None, url=None, resume=True, verbose=1, n_networks=7, thickness='thick')[source]¶
Download and return file names for the Yeo 2011 parcellation.
This function retrieves the so-called yeo deterministic atlases. The provided images are in MNI152 space and have shapes equal to
(256, 256, 256, 1). They contain consecutive integers values from 0 (background) to either 7 or 17 depending on the atlas version considered.For more information on this dataset’s structure, see Cortical Parcellation Estimated by Intrinsic Functional Connectivity[1], and Thomas Yeo et al.[2].
- Parameters:
- data_dir
pathlib.Pathorstror None, optional Path where data should be downloaded. By default, files are downloaded in a
nilearn_datafolder in the home directory of the user. See alsonilearn.datasets.utils.get_data_dirs.- url
stror None, default=None URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data).
- resume
bool, default=True Whether to resume download of a partly-downloaded file.
- verbose
boolorint, default=1 Verbosity level (
0orFalsemeans no message).- n_networks{7, 17}, default = 7
Specify the version of the atlas that is returned:
7 networks parcellation,
17 networks parcellation.
Added in Nilearn 0.12.0.
Changed in Nilearn 0.13.0: The default was changed to 7.
- thickness{“thin”, “thick”}, default = “thick”
Specific the version of the atlas that is returned:
"thick": parcellation fitted to thick cortex segmentations,"thin": parcellation fitted to thin cortex segmentations.
Added in Nilearn 0.12.0.
Changed in Nilearn 0.13.0: The default was changed to “thick”.
- data_dir
- Returns:
- data
sklearn.utils.Bunch Dictionary-like object.
- ‘anat’:
str Path to nifti file containing the anatomy image.
- ‘anat’:
‘maps’: 3D
Nifti1Image. The image contains integer values for each network.- lut
pandas.DataFrame Act as a look up table (lut) with at least columns ‘index’ and ‘name’. Formatted according to ‘dseg.tsv’ format from BIDS.
- lut
- ‘description’
str Description of the dataset.
- ‘description’
- ‘template’
str The standardized space of analysis in which the atlas results are provided. When known it should be a valid template name taken from the spaces described in the BIDS specification.
- ‘template’
- ‘atlas_type’
str Type of atlas. See Probabilistic atlas and Deterministic atlas.
- ‘atlas_type’
- data
Notes
If the dataset files are already present in the user’s Nilearn data directory, this fetcher will not re-download them. To force a fresh download, you can remove the existing dataset folder from your local Nilearn data directory.
For more details on how Nilearn stores datasets.
License: unknown.
References
Examples using nilearn.datasets.fetch_atlas_yeo_2011¶
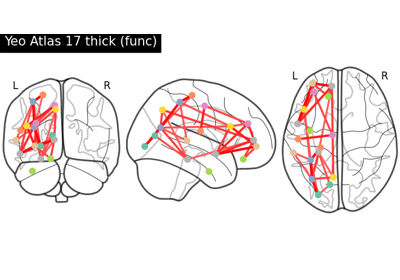
Comparing connectomes on different reference atlases