Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Second-level fMRI model: true positive proportion in clusters¶
This script showcases the so-called “All resolution inference” procedure (Rosenblatt et al.[1]), in which the proportion of true discoveries in arbitrary clusters is estimated. The clusters can be defined from the input image, i.e. in a circular way, as the error control accounts for arbitrary cluster selection.
Fetch dataset¶
We download a list of left vs right button press contrasts from a localizer dataset. Note that we fetch individual t-maps that represent the BOLD activity estimate divided by the uncertainty about this estimate.
from nilearn.datasets import fetch_localizer_contrasts
n_subjects = 16
data = fetch_localizer_contrasts(
["left vs right button press"],
n_subjects,
)
[fetch_localizer_contrasts] Dataset found in
/home/runner/nilearn_data/brainomics_localizer
Estimate second level model¶
We define the input maps and the design matrix for the second level model and fit it.
import pandas as pd
second_level_input = data["cmaps"]
design_matrix = pd.DataFrame(
[1] * len(second_level_input), columns=["intercept"]
)
Model specification and fit
from nilearn.glm.second_level import SecondLevelModel
second_level_model = SecondLevelModel(smoothing_fwhm=8.0, n_jobs=2, verbose=1)
second_level_model = second_level_model.fit(
second_level_input, design_matrix=design_matrix
)
[SecondLevelModel.fit] Fitting second level model. Take a deep breath.
[SecondLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f5d5ff93490>
[SecondLevelModel.fit] Computing mask
[SecondLevelModel.fit] Resampling mask
[SecondLevelModel.fit] Finished fit
[SecondLevelModel.fit]
Computation of second level model done in 0.05 seconds.
To estimate the contrast is very simple. We can just provide the column name of the design matrix.
z_map = second_level_model.compute_contrast(output_type="z_score")
[SecondLevelModel.compute_contrast] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f5d5ff93550>
[SecondLevelModel.compute_contrast] Smoothing images
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
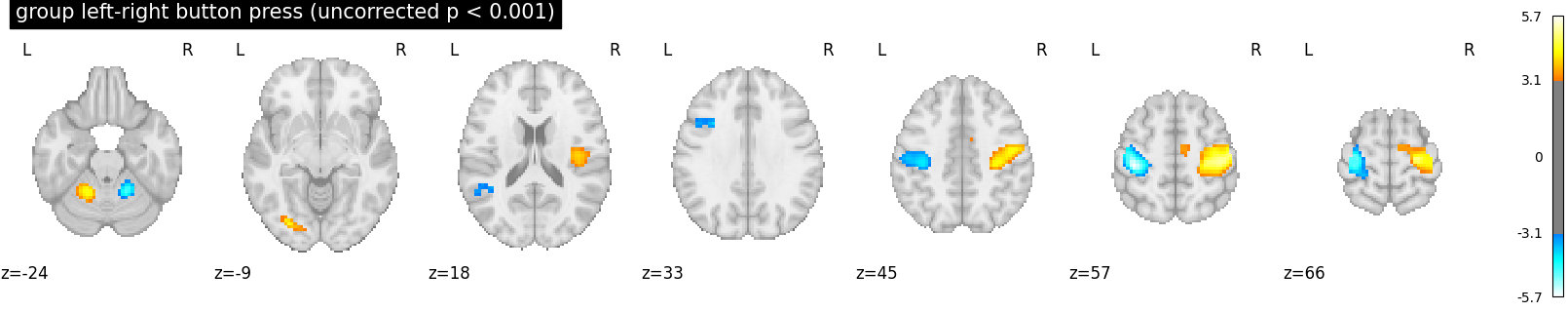
We threshold the second level contrast at uncorrected p < 0.001 and plot
from scipy.stats import norm
from nilearn.glm import cluster_level_inference
from nilearn.plotting import plot_stat_map, show
p_val = 0.001
p001_uncorrected = norm.isf(p_val)
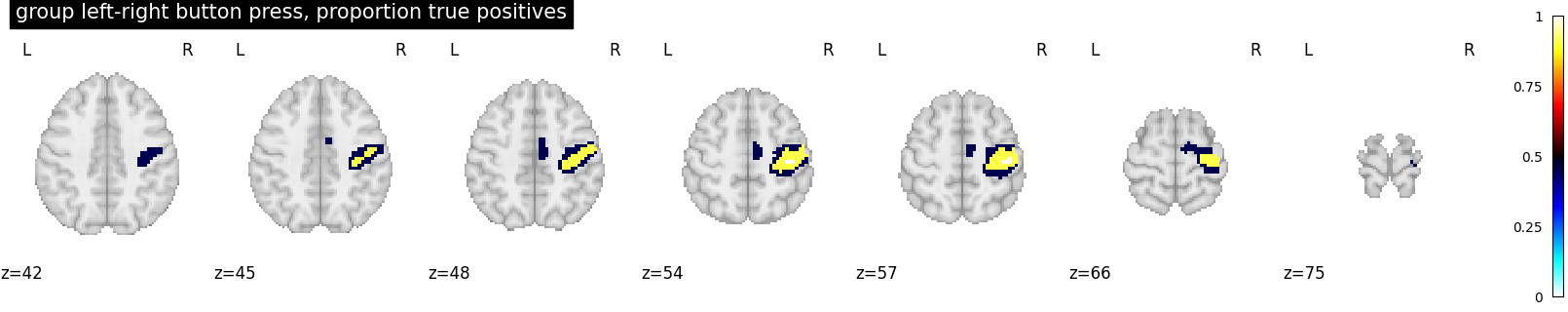
proportion_true_discoveries_img = cluster_level_inference(
z_map, threshold=[2.5, 3.5, 4.5], alpha=0.05
)
data = proportion_true_discoveries_img.get_fdata()
cut_coords = [-24, -9, -18, 33, 45, 57, 66]
plot_stat_map(
proportion_true_discoveries_img,
threshold=0.0,
display_mode="z",
vmax=1,
cmap="inferno",
title="group left-right button press, proportion true positives",
cut_coords=cut_coords,
)
plot_stat_map(
z_map,
threshold=p001_uncorrected,
display_mode="z",
title="group left-right button press (uncorrected p < 0.001)",
cut_coords=cut_coords,
)
show()
References¶
Total running time of the script: (0 minutes 2.670 seconds)
Estimated memory usage: 108 MB