Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Plot Haxby masks¶
Small script to plot the masks of the Haxby dataset.
Load Haxby dataset¶
from nilearn import datasets
from nilearn.plotting import plot_anat, show
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print(
f"First subject anatomical nifti image (3D) is at: {haxby_dataset.anat[0]}"
)
print(
f"First subject functional nifti image (4D) is at: {haxby_dataset.func[0]}"
)
# Build the mean image because we have no anatomic data
from nilearn import image
func_filename = haxby_dataset.func[0]
mean_img = image.mean_img(func_filename)
z_slice = -14
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
First subject anatomical nifti image (3D) is at: /home/runner/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
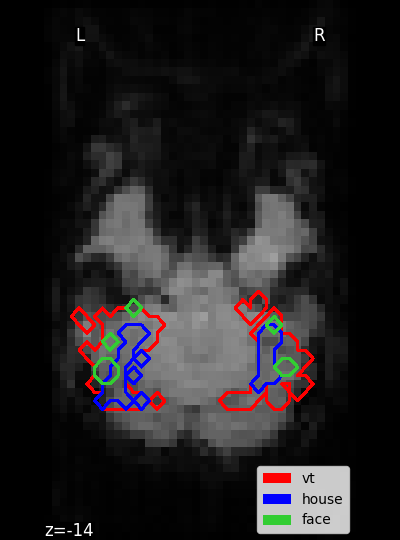
Plot the masks¶
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(4, 5.4), facecolor="k")
display = plot_anat(
mean_img, display_mode="z", cut_coords=[z_slice], figure=fig
)
mask_vt_filename = haxby_dataset.mask_vt[0]
mask_house_filename = haxby_dataset.mask_house[0]
mask_face_filename = haxby_dataset.mask_face[0]
masks = [mask_vt_filename, mask_house_filename, mask_face_filename]
colors = ["red", "blue", "limegreen"]
for mask, color in zip(masks, colors, strict=False):
display.add_contours(
mask,
contours=1,
antialiased=False,
linewidth=4.0,
levels=[0],
colors=[color],
)
# We generate a legend using the trick described on
# https://matplotlib.org/2.0.2/users/legend_guide.html
from matplotlib.patches import Rectangle
p_v = Rectangle((0, 0), 1, 1, fc="red")
p_h = Rectangle((0, 0), 1, 1, fc="blue")
p_f = Rectangle((0, 0), 1, 1, fc="limegreen")
plt.legend([p_v, p_h, p_f], ["vt", "house", "face"], loc="lower right")
show()
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/nilearn/plotting/displays/_axes.py:98: UserWarning:
The following kwargs were not used by contour: 'contours', 'linewidth'
Total running time of the script: (0 minutes 3.633 seconds)
Estimated memory usage: 1002 MB