Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Visualizing a probabilistic atlas: the default mode in the MSDL atlas¶
Visualizing a probabilistic atlas requires visualizing the different maps that compose it.
Here we represent the nodes constituting the default mode network in the MSDL atlas.
The tools that we need to leverage are:
index_imgto retrieve the various maps composing the atlasAdding overlays on an existing brain display, to plot each of these maps
Alternatively, plot_prob_atlas allows
to plot the maps in one step that
with less control over the plot (see below)
Fetching Probabilistic atlas - MSDL atlas¶
from nilearn import datasets
atlas_data = datasets.fetch_atlas_msdl()
atlas_filename = atlas_data.maps
[fetch_atlas_msdl] Dataset created in /home/runner/nilearn_data/msdl_atlas
[fetch_atlas_msdl] Downloading data from
https://team.inria.fr/parietal/files/2015/01/MSDL_rois.zip ...
[fetch_atlas_msdl] ...done. (1 seconds, 0 min)
[fetch_atlas_msdl] Extracting data from
/home/runner/nilearn_data/msdl_atlas/dedd5c670ed1fd457a722109d3c7f958/MSDL_rois.
zip...
[fetch_atlas_msdl] .. done.
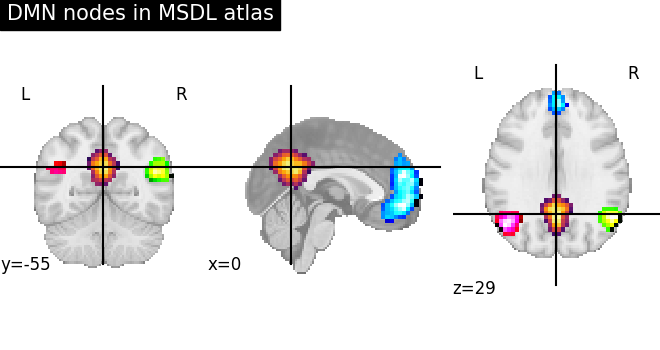
Visualizing a probabilistic atlas with plot_stat_map and add_overlay object¶
from nilearn import image, plotting
# First plot the map for the PCC: index 4 in the atlas
display = plotting.plot_stat_map(
image.index_img(atlas_filename, 4),
colorbar=False,
title="DMN nodes in MSDL atlas",
cmap="inferno",
)
# Now add as an overlay the maps for the ACC and the left and right
# parietal nodes
cmaps = [
plotting.cm.black_blue,
plotting.cm.black_green,
plotting.cm.black_pink,
]
for index, cmap in zip([5, 6, 3], cmaps, strict=False):
display.add_overlay(image.index_img(atlas_filename, index), cmap=cmap)
plotting.show()
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/ma/core.py:2892: UserWarning:
Warning: converting a masked element to nan.
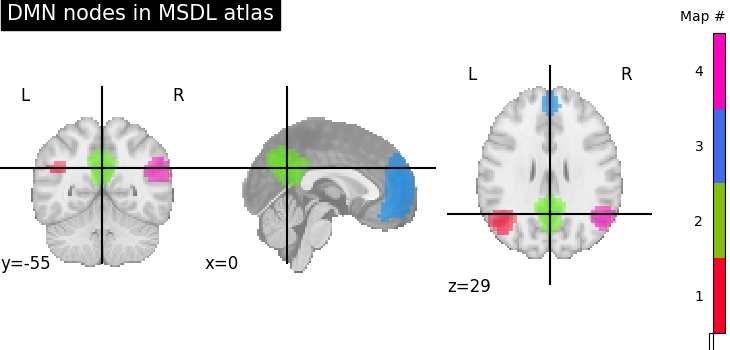
Visualizing a probabilistic atlas with plot_prob_atlas¶
Alternatively, we can create a new 4D-image by selecting
the 3rd, 4th, 5th and 6th (zero-based) probabilistic map from atlas
via index_img
and use plot_prob_atlas (added in version 0.2)
to plot the selected nodes in one step.
Unlike plot_stat_map this works with 4D images
dmn_nodes = image.index_img(atlas_filename, [3, 4, 5, 6])
# Note that dmn_node is now a 4D image
print(dmn_nodes.shape)
(40, 48, 35, 4)
display = plotting.plot_prob_atlas(
dmn_nodes, cut_coords=(0, -55, 29), title="DMN nodes in MSDL atlas"
)
plotting.show()
Total running time of the script: (0 minutes 3.315 seconds)
Estimated memory usage: 107 MB