Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Visualizing Megatrawls Network Matrices from Human Connectome Project¶
This example shows how to fetch network matrices data from HCP beta-release of the Functional Connectivity Megatrawl project.
See fetch_megatrawls_netmats documentation and
the dataset description
for more details.
Fetching the Megatrawls Network matrices¶
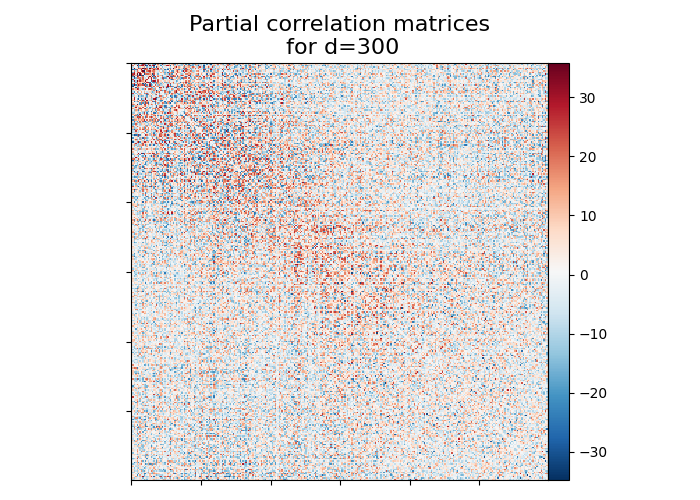
Fetching the partial correlation matrices of dimensionality d=300 with timeseries method ‘eigen regression’
from nilearn.datasets import fetch_megatrawls_netmats
netmats = fetch_megatrawls_netmats(
dimensionality=300,
timeseries="eigen_regression",
matrices="partial_correlation",
)
# Partial correlation matrices array of size (300, 300) are stored in the name
# of 'correlation_matrices'
partial_correlation = netmats.correlation_matrices.to_numpy()
[fetch_megatrawls_netmats] Dataset created in
/home/runner/nilearn_data/Megatrawls
[fetch_megatrawls_netmats] Downloading data from
http://www.nitrc.org/frs/download.php/8037/Megatrawls.tgz ...
[fetch_megatrawls_netmats] ...done. (1 seconds, 0 min)
[fetch_megatrawls_netmats] Extracting data from
/home/runner/nilearn_data/Megatrawls/3f1468dc43a408bf84510b07cee95049/Megatrawls
.tgz...
[fetch_megatrawls_netmats] .. done.
Visualization¶
Import nilearn plotting modules to use its utilities for plotting correlation matrices
from nilearn.plotting import plot_matrix, show
title = "Partial correlation matrices\n for d=300"
display = plot_matrix(partial_correlation, title=title)
show()
Total running time of the script: (0 minutes 1.223 seconds)
Estimated memory usage: 107 MB