Note
Go to the end to download the full example code. or to run this example in your browser via Binder
A short demo of the surface images & maskers¶
copied from the nilearn sandbox discussion, to be transformed into tests & examples
Note
this example is meant to support discussion around a tentative API for surface images in nilearn. This functionality is provided by the nilearn.experimental.surface module; it is still incomplete and subject to change without a deprecation cycle. Please participate in the discussion on GitHub!
from nilearn._utils.helpers import check_matplotlib
check_matplotlib()
import matplotlib.pyplot as plt
import numpy as np
from nilearn.experimental import plotting
from nilearn.experimental.surface import SurfaceMasker, fetch_nki
from nilearn.plotting import plot_matrix
img = fetch_nki()[0]
print(f"NKI image: {img}")
masker = SurfaceMasker()
masked_data = masker.fit_transform(img)
print(f"Masked data shape: {masked_data.shape}")
mean_data = masked_data.mean(axis=0)
mean_img = masker.inverse_transform(mean_data)
print(f"Image mean: {mean_img}")
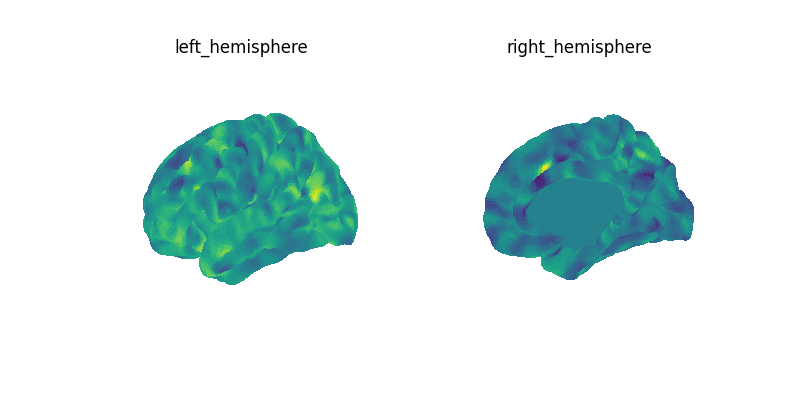
# let's create a figure with all the views for both hemispheres
views = ["lateral", "medial", "dorsal", "ventral", "anterior", "posterior"]
hemispheres = ["left", "right"]
fig, axes = plt.subplots(
len(views),
len(hemispheres),
subplot_kw={"projection": "3d"},
figsize=(4 * len(hemispheres), 4),
)
axes = np.atleast_2d(axes)
for view, ax_row in zip(views, axes):
for ax, hemi in zip(ax_row, hemispheres):
plotting.plot_surf(
surf_map=mean_img,
hemi=hemi,
view=view,
figure=fig,
axes=ax,
title=f"mean image - {hemi} - {view}",
colorbar=False,
cmap="bwr",
symmetric_cmap=True,
bg_on_data=True,
)
fig.set_size_inches(6, 8)
plt.show()
[get_dataset_dir] Dataset found in
/home/runner/nilearn_data/nki_enhanced_surface
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8263/A00033747_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28557312 of 42437884 bytes (67.3%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8262/A00033747_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28819456 of 42406508 bytes (68.0%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8265/A00035072_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28860416 of 42448924 bytes (68.0%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8264/A00035072_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28614656 of 42407636 bytes (67.5%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8267/A00035827_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28098560 of 42421928 bytes (66.2%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8266/A00035827_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28164096 of 42388496 bytes (66.4%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8269/A00035840_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28426240 of 42192604 bytes (67.4%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8268/A00035840_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28286976 of 42210360 bytes (67.0%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8271/A00037112_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28680192 of 42398300 bytes (67.6%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8270/A00037112_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28557312 of 42390948 bytes (67.4%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8273/A00037511_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28819456 of 42308452 bytes (68.1%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8272/A00037511_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28426240 of 42342372 bytes (67.1%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8275/A00038998_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28557312 of 42322216 bytes (67.5%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8274/A00038998_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28237824 of 42300384 bytes (66.8%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8277/A00039391_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28426240 of 42289608 bytes (67.2%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8276/A00039391_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28286976 of 42265584 bytes (66.9%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8279/A00039431_rh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28680192 of 42379532 bytes (67.7%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
[fetch_single_file] Downloading data from
https://www.nitrc.org/frs/download.php/8278/A00039431_lh_preprocessed_fsaverage5
_fwhm6.gii ...
[_chunk_read_] Downloaded 28557312 of 42365696 bytes (67.4%, 0.5s remaining)
[fetch_single_file] ...done. (2 seconds, 0 min)
NKI image: <SurfaceImage (895, 20484)>
Masked data shape: (895, 20484)
Image mean: <SurfaceImage (20484,)>
Connectivity with a surface atlas and SurfaceLabelsMasker¶
from nilearn import connectome
from nilearn.datasets import fetch_atlas_surf_destrieux
from nilearn.experimental.surface import (
SurfaceImage,
SurfaceLabelsMasker,
load_fsaverage,
load_fsaverage_data,
)
# for our plots we will be using the fsaverage sulcal data as background map
fsaverage_sulcal = load_fsaverage_data(data_type="sulcal")
img = fetch_nki()[0]
print(f"NKI image: {img}")
fsaverage = load_fsaverage("fsaverage5")
destrieux = fetch_atlas_surf_destrieux()
labels_img = SurfaceImage(
mesh=fsaverage["pial"],
data={
"left": destrieux["map_left"],
"right": destrieux["map_right"],
},
)
# The labels are stored as bytes for the Destrieux atlas.
# For convenience we decode them to string.
label_names = [x.decode("utf-8") for x in destrieux.labels]
print(f"Destrieux image: {labels_img}")
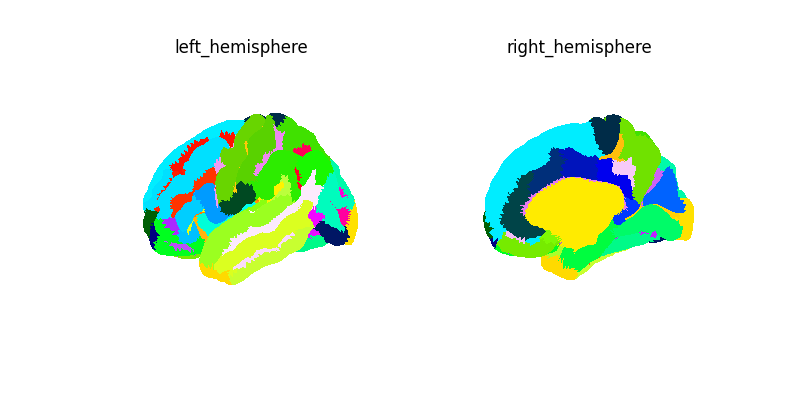
plotting.plot_surf_roi(
roi_map=labels_img,
avg_method="median",
view="lateral",
bg_on_data=True,
bg_map=fsaverage_sulcal,
darkness=0.5,
title="Destrieux atlas",
)
labels_masker = SurfaceLabelsMasker(labels_img, label_names).fit()
report = labels_masker.generate_report()
# This report can be viewed in a notebook
report
# We have several ways to access the report:
# report.open_in_browser()
masked_data = labels_masker.transform(img)
print(f"Masked data shape: {masked_data.shape}")
# or we can save as an html file
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_surface_image_and_maskers"
output_dir.mkdir(exist_ok=True, parents=True)
report.save_as_html(output_dir / "report.html")
[get_dataset_dir] Dataset found in
/home/runner/nilearn_data/nki_enhanced_surface
NKI image: <SurfaceImage (895, 20484)>
[get_dataset_dir] Dataset found in /home/runner/nilearn_data/destrieux_surface
Destrieux image: <SurfaceImage (20484,)>
Masked data shape: (895, 75)
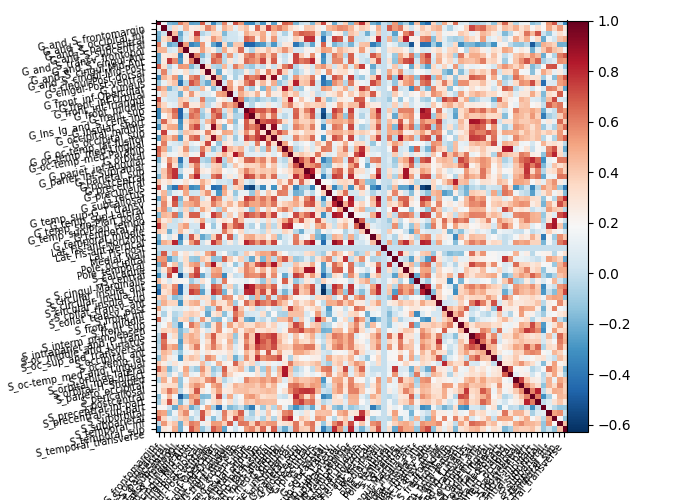
connectome = (
connectome.ConnectivityMeasure(kind="correlation").fit([masked_data]).mean_
)
plot_matrix(connectome, labels=labels_masker.label_names_)
plt.show()
Using the Decoder¶
Now using the appropriate masker we can use a Decoder on surface data just as we do for volume images.
import numpy as np
from nilearn import decoding
img = fetch_nki()[0]
y = np.random.RandomState(0).choice([0, 1], replace=True, size=img.shape[0])
decoder = decoding.Decoder(
mask=SurfaceMasker(),
param_grid={"C": [0.01, 0.1]},
cv=3,
screening_percentile=1,
)
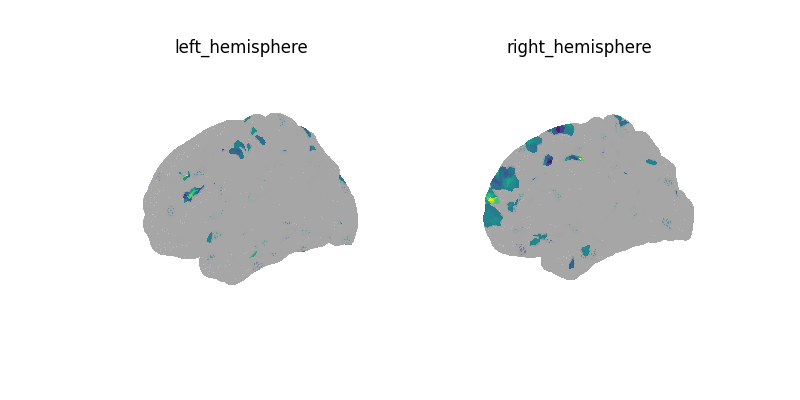
decoder.fit(img, y)
print("CV scores:", decoder.cv_scores_)
plotting.plot_surf(
decoder.coef_img_[0],
threshold=1e-6,
bg_map=fsaverage_sulcal,
bg_on_data=True,
colorbar=True,
cmap="black_red",
vmin=0,
)
plt.show()
[get_dataset_dir] Dataset found in
/home/runner/nilearn_data/nki_enhanced_surface
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/nilearn/decoding/decoder.py:713: UserWarning:
Overriding provided-default estimator parameters with provided masker parameters :
Parameter standardize :
Masker parameter False - overriding estimator parameter True
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in true_divide
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in true_divide
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning:
Features [ 8 36 38 ... 20206 20207 20208] are constant.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.9/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning:
invalid value encountered in true_divide
CV scores: {0: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034], 1: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034]}
Decoding with a scikit-learn Pipeline¶
from sklearn import feature_selection, linear_model, pipeline, preprocessing
img = fetch_nki()[0]
y = np.random.RandomState(0).normal(size=img.shape[0])
decoder = pipeline.make_pipeline(
SurfaceMasker(),
preprocessing.StandardScaler(),
feature_selection.SelectKBest(
score_func=feature_selection.f_regression, k=500
),
linear_model.Ridge(),
)
decoder.fit(img, y)
coef_img = decoder[:-1].inverse_transform(np.atleast_2d(decoder[-1].coef_))
vmax = max([np.absolute(dp).max() for dp in coef_img.data.parts.values()])
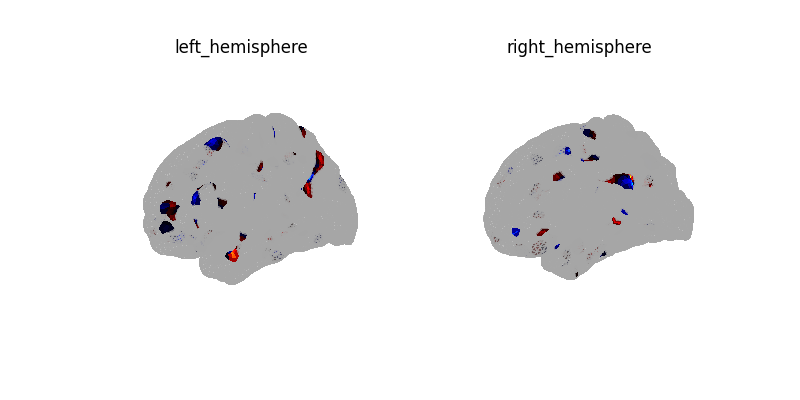
plotting.plot_surf(
coef_img,
cmap="cold_hot",
vmin=-vmax,
vmax=vmax,
threshold=1e-6,
bg_map=fsaverage_sulcal,
bg_on_data=True,
colorbar=True,
)
plt.show()
[get_dataset_dir] Dataset found in
/home/runner/nilearn_data/nki_enhanced_surface
Total running time of the script: (1 minutes 51.312 seconds)
Estimated memory usage: 9533 MB