Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.decoding.Decoder¶
- class nilearn.decoding.Decoder(estimator='svc', mask=None, cv=10, param_grid=None, screening_percentile=20, scoring='roc_auc', smoothing_fwhm=None, standardize=True, target_affine=None, target_shape=None, mask_strategy='background', low_pass=None, high_pass=None, t_r=None, memory=None, memory_level=0, n_jobs=1, verbose=0)[source]¶
A wrapper for popular classification strategies in neuroimaging.
The Decoder object supports classification methods. It implements a model selection scheme that averages the best models within a cross validation loop. The resulting average model is the one used as a classifier. This object also leverages the`NiftiMaskers` to provide a direct interface with the Nifti files on disk.
- Parameters:
- estimator
str, default=’svc’ The estimator to choose among:
"svc":Linear support vector classifierwith L2 penalty.
svc = LinearSVC(penalty="l2", max_iter=1e4)
"svc_l2":Linear support vector classifierwith L2 penalty.
Note
Same as option svc.
"svc_l1":Linear support vector classifierwith L1 penalty.
svc_l1 = LinearSVC(penalty="l1", dual=False, max_iter=1e4)
"logistic":Logistic regressionwith L2 penalty.
logistic = LogisticRegressionCV(penalty="l2", solver="liblinear")
"logistic_l1":Logistic regressionwith L1 penalty.
logistic_l1 = LogisticRegressionCV(penalty="l1", solver="liblinear")
"logistic_l2":Logistic regressionwith L2 penalty
Note
Same as option logistic.
"ridge_classifier":Ridge classifier.
ridge_classifier = RidgeClassifierCV()
"dummy_classifier":Dummy classifier with stratified strategy.
dummy = DummyClassifier(strategy="stratified", random_state=0)
- maskfilename, Nifti1Image, NiftiMasker, MultiNiftiMasker,
SurfaceImageorSurfaceMasker, default=None Mask to be used on data. If an instance of masker is passed, then its mask and parameters will be used. If no mask is given, mask will be computed automatically from provided images by an inbuilt masker with default parameters. Refer to
NiftiMaskerorMultiNiftiMaskerorSurfaceMaskerto check for default parameters.- cvcross-validation generator,
intor None, default=10 A cross-validation generator. See: https://scikit-learn.org/stable/modules/cross_validation.html. If None is passed, cv=10 will be used. It can be an integer, in which case it is the number of folds in a KFold using
StratifiedKFoldwhen groups is None in thefitmethod for this class. If groups is specified butcvis not set to custom CV splitter, default isLeaveOneGroupOut.- param_grid
dictofstrto sequence, or sequence of such, or None, default=None The parameter grid to explore, as a dictionary mapping estimator parameters to sequences of allowed values.
None or an empty dict signifies default parameters.
A sequence of dicts signifies a sequence of grids to search, and is useful to avoid exploring parameter combinations that make no sense or have no effect. See scikit-learn documentation for more information, for example: https://scikit-learn.org/stable/modules/grid_search.html
For DummyClassifier, parameter grid defaults to empty dictionary, class predictions are estimated using default strategy.
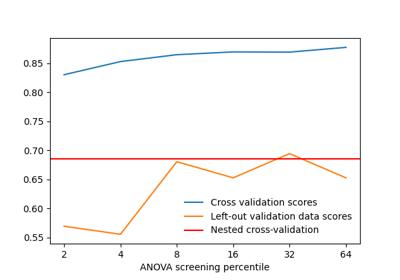
- screening_percentileint, float, in the closed interval [0, 100], or None, default=20
Percentile value for ANOVA univariate feature selection. If
Noneis passed, it will be set to100. A value of100means “keep all features”. This percentile is expressed with respect to the volume of either a standard (MNI152) brain (ifmask_img_is a 3D volume) or a the number of vertices in the mask mesh (ifmask_img_is a SurfaceImage). This means that thescreening_percentileis corrected at runtime by premultiplying it with the ratio of volume of the standard brain to the volume of the mask of the data.Note
If the mask used is too small compared to the total brain volume / surface, then all its elements (voxels / vertices) may be included even for very small
screening_percentile.- scoring
str, callable or None, default=’roc_auc’ The scoring strategy to use. See the scikit-learn documentation at https://scikit-learn.org/stable/modules/model_evaluation.html#the-scoring-parameter-defining-model-evaluation-rules If callable, takes as arguments the fitted estimator, the test data (X_test) and the test target (y_test) if y is not None. e.g. scorer(estimator, X_test, y_test)
For classification, valid entries are: ‘accuracy’, ‘f1’, ‘precision’, ‘recall’ or ‘roc_auc’.
- smoothing_fwhm
floatorintor None, optional. If smoothing_fwhm is not None, it gives the full-width at half maximum in millimeters of the spatial smoothing to apply to the signal.
- standardizeany of: ‘zscore_sample’, ‘zscore’, ‘psc’, True, False or None; default=True
Strategy to standardize the signal:
'zscore_sample': The signal is z-scored. Timeseries are shifted to zero mean and scaled to unit variance. Uses sample std.'zscore': The signal is z-scored. Timeseries are shifted to zero mean and scaled to unit variance. Uses population std by calling defaultnumpy.stdwith N -ddof=0.Deprecated since Nilearn 0.10.1: This option will be removed in Nilearn version 0.14.0. Use
zscore_sampleinstead.'psc': Timeseries are shifted to zero mean value and scaled to percent signal change (as compared to original mean signal).True: The signal is z-scored (same as option zscore). Timeseries are shifted to zero mean and scaled to unit variance.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Truewill be replaced by'zscore_sample'.False: Do not standardize the data.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Falsewill be replaced byNone.
Deprecated since Nilearn 0.10.1: The default will be changed to
'zscore_sample'and'zscore'will be removed in in version 0.14.0.Deprecated since Nilearn 0.15.0dev: The default will be changed to
'zscore_sample'in version 0.15.0.- target_affine3x3 or a 4x4 array-like, or None, default=None
If specified, the image is resampled corresponding to this new affine.
- target_shape
tupleorlistor None, default=None If specified, the image will be resized to match this new shape. len(target_shape) must be equal to 3.
Note
If target_shape is specified, a target_affine of shape (4, 4) must also be given.
- mask_strategy{“background”, “epi”, “whole-brain-template”,”gm-template”, “wm-template”}, optional
The strategy used to compute the mask:
"background": Use this option if your images present a clear homogeneous background. Usesnilearn.masking.compute_background_maskunder the hood."epi": Use this option if your images are raw EPI images. Usesnilearn.masking.compute_epi_mask."whole-brain-template": This will extract the whole-brain part of your data by resampling the MNI152 brain mask for your data’s field of view. Usesnilearn.masking.compute_brain_maskwithmask_type="whole-brain".Note
This option is equivalent to the previous ‘template’ option which is now deprecated.
"gm-template": This will extract the gray matter part of your data by resampling the corresponding MNI152 template for your data’s field of view. Usesnilearn.masking.compute_brain_maskwithmask_type="gm".Added in Nilearn 0.8.1.
"wm-template": This will extract the white matter part of your data by resampling the corresponding MNI152 template for your data’s field of view. Usesnilearn.masking.compute_brain_maskwithmask_type="wm".Added in Nilearn 0.8.1.
Note
This parameter will be ignored if a mask image is provided.
Note
Depending on this value, the mask will be computed from
nilearn.masking.compute_background_mask,nilearn.masking.compute_epi_mask, ornilearn.masking.compute_brain_mask.Default=’background’.
- low_pass
floatorintor None, default=None Low cutoff frequency in Hertz. If specified, signals above this frequency will be filtered out. If None, no low-pass filtering will be performed.
- high_pass
floatorintor None, default=None High cutoff frequency in Hertz. If specified, signals below this frequency will be filtered out.
- t_r
floatorintor None, default=None Repetition time, in seconds (sampling period). Set to None if not provided.
- memoryNone, instance of
joblib.Memory,str, orpathlib.Path, default=None Used to cache the masking process. By default, no caching is done. If a
stris given, it is the path to the caching directory.- memory_level
int, default=0 Rough estimator of the amount of memory used by caching. Higher value means more memory for caching. Zero means no caching.
- n_jobs
int, default=1 The number of CPUs to use to do the computation. -1 means ‘all CPUs’.
- verbose
boolorint, default=0 Verbosity level (
0orFalsemeans no message).
- estimator
- Attributes:
- coef_numpy.ndarray, shape=(n_classes, n_features)
Contains the mean of the models weight vector across fold for each class. Returns None for Dummy estimators.
- coef_img_
dictof Nifti1Image Dictionary containing
coef_with class names as keys, andcoef_transformed in Nifti1Images as values. In the case of a regression, it contains a single Nifti1Image at the key ‘beta’. Ignored if Dummy estimators are provided.- cv_
listof pairs of lists List of the (n_folds,) folds. For the corresponding fold, each pair is composed of two lists of indices, one for the train samples and one for the test samples.
- cv_params_
dictoflist Best point in the parameter grid for each tested fold in the inner cross validation loop. The grid is empty when Dummy estimators are provided.
Note
If the estimator used its built-in cross-validation, this will include an additional key for the single best value estimated by the built-in cross-validation (‘best_C’ for LogisticRegressionCV and ‘best_alpha’ for RidgeCV/RidgeClassifierCV/LassoCV), in addition to the input list of values.
- cv_scores_
dict, (classes, n_folds) Scores (misclassification) for each parameter, and on each fold
- dummy_output_ndarray, shape=(n_classes, 2) or shape=(1, 1) for regression
Contains dummy estimator attributes after class predictions using strategies of
sklearn.dummy.DummyClassifier(class_prior) andsklearn.dummy.DummyRegressor(constant) from scikit-learn. This attribute is necessary for estimating class predictions after fit. Returns None if non-dummy estimators are provided.- estimator_Estimator object used during decoding.
- intercept_ndarray, shape (nclasses,)
Intercept (also known as bias) added to the decision function. Ignored if Dummy estimators are provided.
- mask_img_Nifti1Image or
SurfaceImage Mask computed by the masker object.
- masker_instance of NiftiMasker, MultiNiftiMasker, or SurfaceMasker
The masker used to mask the data.
- memory_joblib memory cache
- n_elements_
int The number of voxels or vertices in the mask.
Added in Nilearn 0.12.1.
- n_outputs_
int Number of outputs (column-wise)
- scorer_function
Scorer function used on the held out data to choose the best parameters for the model.
- screening_percentile_
float Percentile value for ANOVA univariate feature selection. A value of 100 means ‘keep all features’. This percentile is expressed with respect to the volume of either a standard (MNI152) brain (if mask_img is a 3D volume) or a the number of vertices in the mask mesh (if mask_img is a SurfaceImage).
- std_coef_numpy.ndarray, shape=(n_classes, n_features)
Contains the standard deviation of the models weight vector across fold for each class. Note that folds are not independent, see https://scikit-learn.org/stable/modules/cross_validation.html#cross-validation-iterators-for-grouped-data Ignored if Dummy estimators are provided.
- std_coef_img_
dictof Nifti1Image Dictionary containing std_coef_ with class names as keys, and coef_ transformed in Nifti1Image as values. In the case of a regression, it contains a single Nifti1Image at the key ‘beta’. Ignored if Dummy estimators are provided.
- classes_ndarray of labels (n_classes_)
Labels of the classes
- n_classes_int
number of classes
See also
nilearn.decoding.DecoderRegressorregression strategies for Neuro-imaging,
nilearn.decoding.FREMClassifierState of the art classification pipeline for Neuroimaging
nilearn.decoding.SpaceNetClassifierGraph-Net and TV-L1 priors/penalties
- __init__(estimator='svc', mask=None, cv=10, param_grid=None, screening_percentile=20, scoring='roc_auc', smoothing_fwhm=None, standardize=True, target_affine=None, target_shape=None, mask_strategy='background', low_pass=None, high_pass=None, t_r=None, memory=None, memory_level=0, n_jobs=1, verbose=0)[source]¶
- decision_function(X)[source]¶
Predict class labels for samples in X.
- Parameters:
- XNiimg-like,
SurfaceImage,listof Niimg-like objects orlistofSurfaceImage, or {array-like, sparse matrix}, shape = (n_samples, n_features) See Input and output: neuroimaging data representation. Data on prediction is to be made. If this is a list, the affine is considered the same for all.
- XNiimg-like,
- Returns:
- y_pred
numpy.ndarray, shape (n_samples,) Predicted class label per sample.
- y_pred
- fit(X, y, groups=None)[source]¶
Fit the decoder (learner).
- Parameters:
- Xlist of Niimg-like or
SurfaceImageobjects See Input and output: neuroimaging data representation. Data on which model is to be fitted. If this is a list, the affine is considered the same for all.
- ynumpy.ndarray of shape=(n_samples) or list of length n_samples
The dependent variable (age, sex, IQ, yes/no, etc.). Target variable to predict. Must have exactly as many samples as the input images.
- groupsNone, default=None
Group labels for the samples used while splitting the dataset into train/test set.
Note that this parameter must be specified in some scikit-learn cross-validation generators to calculate the number of splits, for example sklearn.model_selection.LeaveOneGroupOut or sklearn.model_selection.LeavePGroupsOut.
For more details see https://scikit-learn.org/stable/modules/cross_validation.html#cross-validation-iterators-for-grouped-data
- Xlist of Niimg-like or
- get_metadata_routing()¶
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- predict(X)[source]¶
Predict a label for all X vectors indexed by the first axis.
- Parameters:
- XNiimg-like,
SurfaceImage,listof Niimg-like objects orlistofSurfaceImage, or {array-like, sparse matrix}, shape = (n_samples, n_features) See Input and output: neuroimaging data representation. Data on which prediction is to be made.
- XNiimg-like,
- Returns:
- array, shape=(n_samples,) if n_classes == 2 else (n_samples, n_classes)
Confidence scores per (sample, class) combination. In the binary case, confidence score for self.classes_[1] where >0 means this class would be predicted.
- score(X, y, *args)[source]¶
Compute the prediction score using the scoring metric defined by the scoring attribute.
- Parameters:
- XNiimg-like,
SurfaceImage,listof Niimg-like objects orlistofSurfaceImage, or {array-like, sparse matrix}, shape = (n_samples, n_features) See Input and output: neuroimaging data representation. Data on which prediction is to be made.
- y
numpy.ndarray Target values.
- argsOptional arguments that can be passed to
scoring metrics. Example: sample_weight.
- XNiimg-like,
- Returns:
- scorefloat
Prediction score.
- set_fit_request(*, groups='$UNCHANGED$')¶
Configure whether metadata should be requested to be passed to the
fitmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- groupsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
groupsparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using nilearn.decoding.Decoder¶
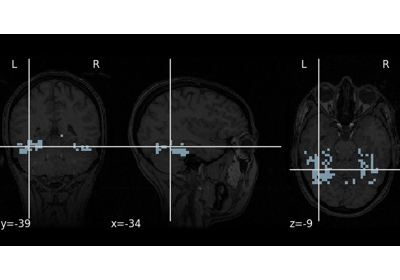
Decoding of a dataset after GLM fit for signal extraction
Decoding with ANOVA + SVM: face vs house in the Haxby dataset
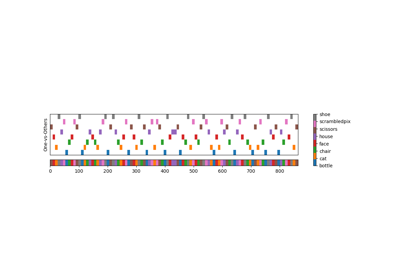
Different classifiers in decoding the Haxby dataset
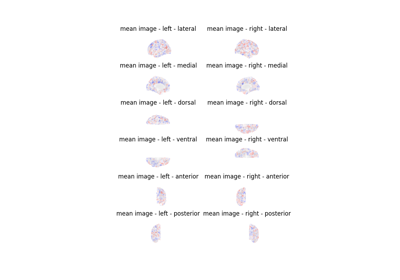
ROI-based decoding analysis in Haxby et al. dataset