Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.load_fsaverage¶
- nilearn.datasets.load_fsaverage(mesh='fsaverage5', data_dir=None)[source]¶
Load fsaverage for both hemispheres as PolyMesh objects.
Added in Nilearn 0.11.0.
- Parameters:
- mesh
str, default=’fsaverage5’ Which mesh to fetch. Should be one of the following values:
"fsaverage3": the low-resolution fsaverage3 mesh (642 nodes)"fsaverage4": the low-resolution fsaverage4 mesh (2562 nodes)"fsaverage5": the low-resolution fsaverage5 mesh (10242 nodes)"fsaverage6": the medium-resolution fsaverage6 mesh (40962 nodes)"fsaverage7": same as “fsaverage”"fsaverage": the high-resolution fsaverage mesh (163842 nodes)
Note
The high-resolution fsaverage will result in more computation time and memory usage
- data_dir
pathlib.Pathorstror None, optional Path where data should be downloaded. By default, files are downloaded in a
nilearn_datafolder in the home directory of the user. See alsonilearn.datasets.utils.get_data_dirs.
- mesh
- Returns:
- data
sklearn.utils.Bunch - Dictionary-like object, the interest attributes are :
'description': description of the dataset'pial': Polymesh for pial surface for left and right hemispheres'white_matter': Polymesh for white matter surfacefor left and right hemispheres
'inflated': Polymesh for inglated surfacefor left and right hemispheres
'sphere': Polymesh for spherical surfacefor left and right hemispheres
'flat': Polymesh for flattened surfacefor left and right hemispheres
- data
Examples using nilearn.datasets.load_fsaverage¶
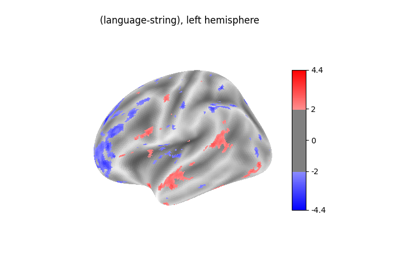
Surface-based dataset first and second level analysis of a dataset