Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.plotting.displays.XSlicer¶
- class nilearn.plotting.displays.XSlicer(cut_coords, axes=None, black_bg=False, brain_color=(0.5, 0.5, 0.5), **kwargs)[source]¶
The
XSlicerclass enables sagittal visualization with plotting functions of Nilearn likenilearn.plotting.plot_img.This visualization mode can be activated by setting
display_mode='x':from nilearn.datasets import load_mni152_template from nilearn.plotting import plot_img img = load_mni152_template() # display is an instance of the XSlicer class display = plot_img(img, display_mode="x")
- Parameters:
- cut_coords
int, sequence offloatorintor None The number of cuts to perform or the list of cut positions in the direction ‘x’. If
None, 7 cut positions are calculated automatically.- axes
matplotlib.axes.Axes, or 4tupleoffloat: (xmin, ymin, width, height), default=None The axes, or the coordinates, in matplotlib figure space, of the axes used to display the plot. If None, the complete figure is used.
- black_bg
bool, or “auto”, optional If True, the background of the image is set to be black. If you wish to save figures with a black background, you will need to pass facecolor=”k”, edgecolor=”k” to
matplotlib.pyplot.savefig.- brain_color
tuple, default=(0.5, 0.5, 0.5) The brain color to use as the background color (e.g., for transparent colorbars).
- kwargs
dict Extra keyword arguments are passed to
CutAxesused for plotting in each direction.
- cut_coords
- Attributes:
See also
nilearn.plotting.displays.YSlicerCoronal view
nilearn.plotting.displays.ZSlicerAxial view
- add_contours(img, threshold=1e-06, filled=False, **kwargs)[source]¶
Contour a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. Provides image to plot.
- threshold
intorfloatorNone, default=1e-6 Threshold to apply:
If
Noneis given, the maps are not thresholded.- If number is given, it must be non-negative. The specified
value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent.
- filled
bool, default=False If
filled=True, contours are displayed with color fillings.- kwargs
dict Extra keyword arguments are passed to function
contour, or functioncontourf. Useful, arguments are typical “levels”, which is a list of values to use for plotting a contour or contour fillings (iffilled=True), and “colors”, which is one color or a list of colors for these contours.
- Raises:
- ValueError
if the specified threshold is a negative number
Notes
If colors are not specified, default coloring choices (from matplotlib) for contours and contour_fillings can be different.
- add_edges(img, color='r')[source]¶
Plot the edges of a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. The 3D map to be plotted. If it is a masked array, only the non-masked part will be plotted.
- colormatplotlib color:
stror (r, g, b) value, default=’r’ The color used to display the edge map.
- add_markers(marker_coords, marker_color='r', marker_size=30, **kwargs)[source]¶
Add markers to the plot.
- Parameters:
- marker_coords
ndarrayof shape(n_markers, 3) Coordinates of the markers to plot. For each slice, only markers that are 2 millimeters away from the slice are plotted.
- marker_colorpyplot compatible color or
listof shape(n_markers,), default=’r’ List of colors for each marker that can be string or matplotlib colors.
- marker_size
floatorlistoffloatof shape(n_markers,), default=30 Size in pixel for each marker.
- kwargs
dict Extra keyword arguments are passed to
matplotlib.pyplot.scatter.
- marker_coords
- add_overlay(img, threshold=1e-06, colorbar=False, cbar_tick_format='%.2g', cbar_vmin=None, cbar_vmax=None, transparency=None, transparency_range=None, **kwargs)[source]¶
Plot a 3D map in all the views.
- Parameters:
- imgNiimg-like object
See Input and output: neuroimaging data representation. If it is a masked array, only the non-masked part will be plotted.
- threshold
intorfloatorNone, default=1e-6 Threshold to apply:
If
Noneis given, the maps are not thresholded.- If number is given, it must be non-negative. The specified
value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent.
- colorbar
bool, optional If True, display a colorbar next to the plots. Default=False.
- cbar_tick_formatstr, default=”%.2g” (scientific notation)
Controls how to format the tick labels of the colorbar. Ex: use “%i” to display as integers.
- cbar_vmin
float, default=None Minimal value for the colorbar. If None, the minimal value is computed based on the data.
- cbar_vmax
float, default=None Maximal value for the colorbar. If None, the maximal value is computed based on the data.
- transparency
floatbetween 0 and 1, or a Niimg-Like object, or None, default = None Value to be passed as alpha value to
imshow. ifNoneis passed, it will be set to 1. If an image is passed, voxel-wise alpha blending will be applied, by relying on the absolute value oftransparencyat each voxel.Added in Nilearn 0.12.0.
- transparency_range
tupleorlistof 2 non-negative numbers, or None, default = None When an image is passed to
transparency, this determines the range of values in the image to use for transparency (alpha blending). For example withtransparency_range = [1.96, 3], any voxel / vertex (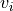 ):
):with a value between between -1.96 and 1.96, would be fully transparent (alpha = 0),
with a value less than -3 or greater than 3, would be fully opaque (alpha = 1),
with a value in the intervals
[-3.0, -1.96]or[1.96, 3.0], would have an alpha_i value scaled linearly between 0 and 1 :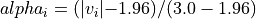 .
.
This parameter will be ignored unless an image is passed as
transparency. The first number must be greater than 0 and less than the second one. ifNoneis passed, this will be set to[0, max(abs(transparency))].Added in Nilearn 0.12.0.
- kwargs
dict Extra keyword arguments are passed to function
imshow.
- Raises:
- ValueError
if the specified threshold is a negative number
- annotate(left_right=True, positions=True, scalebar=False, size=12, scale_size=5.0, scale_units='cm', scale_loc=4, decimals=0, **kwargs)[source]¶
Add annotations to the plot.
- Parameters:
- left_right
bool, default=True If
True, annotations indicating which side is left and which side is right are drawn.- positions
bool, default=True If
True, annotations indicating the positions of the cuts are drawn.- scalebar
bool, default=False If
True, cuts are annotated with a reference scale bar. For finer control of the scale bar, please check out thedraw_scale_barmethod on the axes in “axes” attribute of this object.- size
int, default=12 The size of the text used.
- scale_size
intorfloat, default=5.0 The length of the scalebar, in units of
scale_units.- scale_units{‘cm’, ‘mm’}, default=’cm’
The units for the
scalebar.- scale_loc
int, default=4 The positioning for the scalebar. Valid location codes are:
1: “upper right”
2: “upper left”
3: “lower left”
4: “lower right”
5: “right”
6: “center left”
7: “center right”
8: “lower center”
9: “upper center”
10: “center”
- decimals
int, default=0 Number of decimal places on slice position annotation. If zero, the slice position is integer without decimal point.
- kwargs
dict Extra keyword arguments are passed to matplotlib’s text function.
- left_right
- property black_bg¶
Return black background.
- property brain_color¶
Return brain color.
- draw_cross(cut_coords=None, **kwargs)[source]¶
Draw a crossbar on the plot to show where the cut is performed.
Not implemented for this slicer.
- Parameters:
- cut_coords
int, sequence offloatorintor None, default=None The list of positions of the crosses to draw in the direction of this slicer. If
Noneis passed, the this slicer’s cut coordinates are used.- kwargs
dict Extra keyword arguments are passed to function
matplotlib.pyplot.axhline.
- cut_coords
- classmethod find_cut_coords(img=None, threshold=None, cut_coords=None)[source]¶
Find world coordinates of cut positions compatible with this slicer.
- Parameters:
- img3D
Nifti1Image, default=None The brain image.
- threshold
float, default=None The lower threshold to the positive activation. If
None, the activation threshold is computed using the 80% percentile of the absolute value of the map.- cut_coords
int, sequence offloatorintor None, default=None The number of cuts to perform or the list of cut positions in the direction of this slicer. If
Noneis given, the cuts are calculated automatically.
- img3D
- Returns:
- classmethod init_with_figure(img, threshold=None, cut_coords=None, figure=None, axes=None, black_bg=False, leave_space=False, colorbar=False, brain_color=(0.5, 0.5, 0.5), **kwargs)[source]¶
Initialize the slicer with an image.
- Parameters:
- imgNiimg-like object
- threshold
intorfloat, None, or ‘auto’, optional If None is given, the image is not thresholded. If number is given, it must be non-negative. The specified value is used to threshold the image: values below the threshold (in absolute value) are plotted as transparent. If “auto” is given, the threshold is determined based on the score obtained using percentile value “80%” on the absolute value of the image data.
- cut_coordsNone, allowed types depend on the
display_mode, optional The world coordinates of the point where the cut is performed.
If
display_modeis'ortho'or'tiled', this must be a 3-sequence offloatorint:(x, y, z).If
display_modeis'xz','yz'or'yx', this must be a 2-sequence offloatorint:(x, z),(y, z)or(x, y).If
display_modeis"x","y", or"z", this can be:If
display_modeis'mosaic', this can be:an
intin which case it specifies the number of cuts to perform in each direction"x","y","z".a 3-sequence of
floatorintin which case it specifies the number of cuts to perform in each direction"x","y","z"separately.dict<str: 1Dndarray> in which case keys are the directions (‘x’, ‘y’, ‘z’) and the values are sequences holding the cut coordinates.
If
Noneis given, the cuts are calculated automatically.
- figure
matplotlib.figure.Figure Figure to be used for plots.
- axes
matplotlib.axes.Axes, or 4tupleoffloat: (xmin, ymin, width, height), default=None The axes, or the coordinates, in matplotlib figure space, of the axes used to display the plot. If None, the complete figure is used. default=None The axes that will be subdivided in 3.
- black_bg
bool, or “auto”, optional If True, the background of the image is set to be black. If you wish to save figures with a black background, you will need to pass facecolor=”k”, edgecolor=”k” to
matplotlib.pyplot.savefig. default=False- leave_space
bool, default=False If
True, leave space between the plots.- colorbar
bool, optional If True, display a colorbar next to the plots. Default=False.
- brain_color
tuple, default=(0.5, 0.5, 0.5) The brain color to use as the background color (e.g., for transparent colorbars).
- kwargs
dict Extra keyword arguments are passed to
CutAxesused for plotting in each direction.
- Raises:
- ValueError
if the specified threshold is a negative number
- savefig(filename, dpi=None, **kwargs)[source]¶
Save the figure to a file.
- Parameters:
- filename
str The file name to save to. Its extension determines the file type, typically ‘.png’, ‘.svg’ or ‘.pdf’.
- dpiNone or scalar, default=None
The resolution in dots per inch.
- kwargs
dict Extra keyword arguments are passed to
matplotlib.pyplot.savefig.
- filename
- title(text, x=0.01, y=0.99, size=15, color=None, bgcolor=None, alpha=1, **kwargs)[source]¶
Write a title to the view.
- Parameters:
- text
str The text of the title.
- x
float, default=0.01 The horizontal position of the title on the frame in fraction of the frame width.
- y
float, default=0.99 The vertical position of the title on the frame in fraction of the frame height.
- size
int, default=15 The size of the title text.
- colormatplotlib color specifier, default=None
The color of the font of the title.
- bgcolormatplotlib color specifier, default=None
The color of the background of the title.
- alpha
float, default=1 The alpha value for the background.
- kwargs
dict Extra keyword arguments are passed to matplotlib’s text function.
- text