Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_msdl¶
- nilearn.datasets.fetch_atlas_msdl(data_dir=None, url=None, resume=True, verbose=1)[source]¶
Download and load the MSDL brain Probabilistic atlas.
It can be downloaded at Spatially Constrained Parcellation[1], and cited using Varoquaux et al.[2]. See also Varoquaux and Craddock[3] for more information.
- Parameters:
- data_dir
pathlib.Pathorstror None, optional Path where data should be downloaded. By default, files are downloaded in a
nilearn_datafolder in the home directory of the user. See alsonilearn.datasets.utils.get_data_dirs.- url
stror None, default=None URL of file to download. Override download URL. Used for test only (or if you setup a mirror of the data).
- resume
bool, default=True Whether to resume download of a partly-downloaded file.
- verbose
boolorint, default=1 Verbosity level (
0orFalsemeans no message).
- data_dir
- Returns:
- data
sklearn.utils.Bunch Dictionary-like object, the interest attributes are :
- ‘maps’:
str path to nifti file containing the Probabilistic atlas image (shape is equal to
(40, 48, 35, 39)).
- ‘maps’:
- ‘description’
str Description of the dataset.
- ‘description’
- ‘atlas_type’
str Type of atlas. See Probabilistic atlas and Deterministic atlas.
- ‘atlas_type’
- ‘template’
str The standardized space of analysis in which the atlas results are provided. When known it should be a valid template name taken from the spaces described in the BIDS specification.
- ‘template’
- data
References
Examples using nilearn.datasets.fetch_atlas_msdl¶
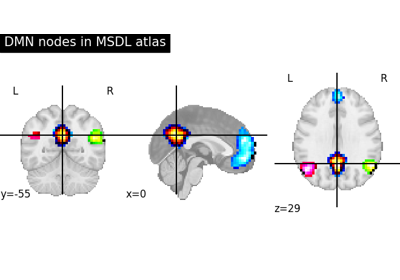
Visualizing a probabilistic atlas: the default mode in the MSDL atlas
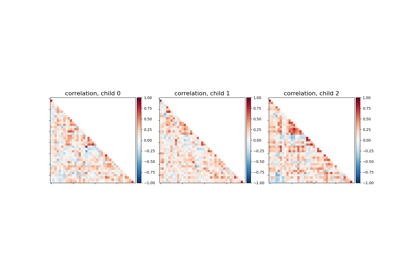
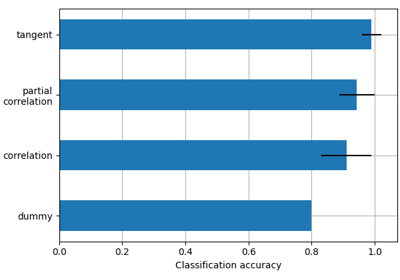
Classification of age groups using functional connectivity
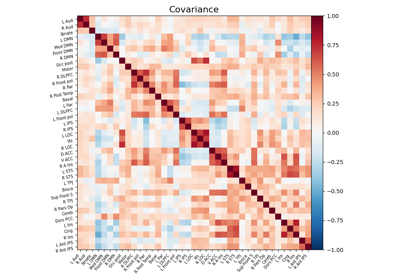
Computing a connectome with sparse inverse covariance
Extracting signals of a probabilistic atlas of functional regions
Group Sparse inverse covariance for multi-subject connectome