Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.datasets.fetch_atlas_harvard_oxford¶
- nilearn.datasets.fetch_atlas_harvard_oxford(atlas_name, data_dir=None, symmetric_split=False, resume=True, verbose=1)[source]¶
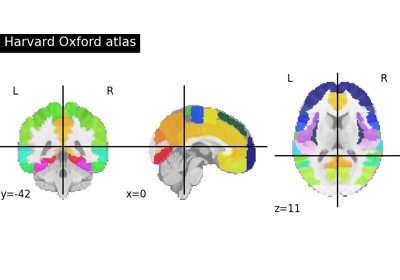
Load Harvard-Oxford parcellations from FSL.
This function downloads Harvard Oxford atlas packaged from FSL 5.0 and stores atlases in NILEARN_DATA folder in home directory.
This function can also load Harvard Oxford atlas from your local directory specified by your FSL installed path given in data_dir argument. See documentation for details.
Note
For atlases ‘cort-prob-1mm’, ‘cort-prob-2mm’, ‘cortl-prob-1mm’, ‘cortl-prob-2mm’, ‘sub-prob-1mm’, and ‘sub-prob-2mm’, the function returns a Probabilistic atlas, and the
Nifti1Imagereturned is 4D, with shape(182, 218, 182, 48). For deterministic atlases, theNifti1Imagereturned is 3D, with shape(182, 218, 182)and 48 regions (+ background).- Parameters:
- atlas_name
str Name of atlas to load. Can be: “cort-maxprob-thr0-1mm”, “cort-maxprob-thr0-2mm”, “cort-maxprob-thr25-1mm”, “cort-maxprob-thr25-2mm”, “cort-maxprob-thr50-1mm”, “cort-maxprob-thr50-2mm”, “cort-prob-1mm”, “cort-prob-2mm”, “cortl-maxprob-thr0-1mm”, “cortl-maxprob-thr0-2mm”, “cortl-maxprob-thr25-1mm”, “cortl-maxprob-thr25-2mm”, “cortl-maxprob-thr50-1mm”, “cortl-maxprob-thr50-2mm”, “cortl-prob-1mm”, “cortl-prob-2mm”, “sub-maxprob-thr0-1mm”, “sub-maxprob-thr0-2mm”, “sub-maxprob-thr25-1mm”, “sub-maxprob-thr25-2mm”, “sub-maxprob-thr50-1mm”, “sub-maxprob-thr50-2mm”, “sub-prob-1mm”, “sub-prob-2mm”.
- data_dir
pathlib.Pathorstror None, optional Path where data should be downloaded. By default, files are downloaded in a
nilearn_datafolder in the home directory of the user. See alsonilearn.datasets.utils.get_data_dirs. Optionally, it can also be a FSL installation directory (which is dependent on your installation). Example, if FSL is installed in/usr/share/fsl/then specifying as ‘/usr/share/’ can get you the Harvard Oxford atlas from your installed directory. Since we mimic the same root directory as FSL to load it easily from your installation.- symmetric_split
bool, default=False If
True, lateralized atlases of cort or sub with maxprob will be returned. For subcortical types (sub-maxprob), we split every symmetric region in left and right parts. Effectively doubles the number of regions.Note
Not implemented for full Probabilistic atlas (-prob- atlases).
- resume
bool, default=True Whether to resume download of a partly-downloaded file.
- verbose
boolorint, default=1 Verbosity level (
0orFalsemeans no message).
- atlas_name
- Returns:
- data
sklearn.utils.Bunch Dictionary-like object, keys are:
- ‘maps’:
str path to nifti file containing the atlas
Nifti1Image. It is a 4D image if a Probabilistic atlas is requested, and a 3D image if a maximum probability atlas is requested. In the latter case, the image contains integer values which can be interpreted as the indices in the list of labels.Note
For some atlases, it can be the case that some regions are empty. In this case, no voxels in the map are assigned to these regions. So the number of unique values in the map can be strictly smaller than the number of region names in
labels.
- ‘maps’:
‘filename’: Same as ‘maps’, kept for backward compatibility only.
- ‘description’
str Description of the dataset.
- ‘description’
- lut
pandas.DataFrame Act as a look up table (lut) with at least columns ‘index’ and ‘name’. Formatted according to ‘dseg.tsv’ format from BIDS. Only for deterministic version of the atlas.
- lut
- ‘template’
str The standardized space of analysis in which the atlas results are provided. When known it should be a valid template name taken from the spaces described in the BIDS specification.
- ‘template’
- ‘atlas_type’
str Type of atlas. See Probabilistic atlas and Deterministic atlas.
- ‘atlas_type’
- data
See also
Examples using nilearn.datasets.fetch_atlas_harvard_oxford¶
Extracting signals from brain regions using the NiftiLabelsMasker