Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Decoding with ANOVA + SVM: face vs house in the Haxby dataset¶
This example does a simple but efficient decoding on the Haxby dataset: using a feature selection, followed by an SVM.
Retrieve the files of the Haxby dataset¶
from nilearn import datasets
# By default 2nd subject will be fetched
haxby_dataset = datasets.fetch_haxby()
func_img = haxby_dataset.func[0]
# print basic information on the dataset
print(f"Mask nifti image (3D) is located at: {haxby_dataset.mask}")
print(f"Functional nifti image (4D) is located at: {func_img}")
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
Mask nifti image (3D) is located at: /home/runner/nilearn_data/haxby2001/mask.nii.gz
Functional nifti image (4D) is located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Load the behavioral data¶
import pandas as pd
# Load target information as string and give a numerical identifier to each
behavioral = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
conditions = behavioral["labels"]
# Restrict the analysis to faces and places
from nilearn.image import index_img
condition_mask = behavioral["labels"].isin(["face", "house"])
conditions = conditions[condition_mask]
func_img = index_img(func_img, condition_mask)
# Confirm that we now have 2 conditions
print(conditions.unique())
# The number of the run is stored in the CSV file giving the behavioral data.
# We have to apply our run mask, to select only faces and houses.
run_label = behavioral["chunks"][condition_mask]
['face' 'house']
ANOVA pipeline with Decoder object¶
Nilearn Decoder object aims to provide smooth user experience by acting as a pipeline of several tasks: preprocessing with NiftiMasker, reducing dimension by selecting only relevant features with ANOVA – a classical univariate feature selection based on F-test, and then decoding with different types of estimators (in this example is Support Vector Machine with a linear kernel) on nested cross-validation.
Fit the decoder and predict¶
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0cd65960>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_anova_svm.py:73: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0cd65960>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 5
[Decoder.fit] Corrected screening-percentile: 4.79274
[Parallel(n_jobs=1)]: Done 10 out of 10 | elapsed: 1.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0cd65960>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Obtain prediction scores via cross validation¶
Define the cross-validation scheme used for validation. Here we use a LeaveOneGroupOut cross-validation on the run group which corresponds to a leave a run out scheme, then pass the cross-validator object to the cv parameter of decoder.leave-one-session-out. For more details please take a look at: A introduction tutorial to fMRI decoding.
from sklearn.model_selection import LeaveOneGroupOut
cv = LeaveOneGroupOut()
decoder = Decoder(
estimator="svc",
mask=mask_img,
standardize="zscore_sample",
screening_percentile=5,
scoring="accuracy",
cv=cv,
verbose=1,
)
# Compute the prediction accuracy for the different folds (i.e. run)
decoder.fit(func_img, conditions, groups=run_label)
# Print the CV scores
print(decoder.cv_scores_["face"])
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0cd65960>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_anova_svm.py:99: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0cd65960>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 5
[Decoder.fit] Corrected screening-percentile: 4.79274
[Parallel(n_jobs=1)]: Done 12 out of 12 | elapsed: 1.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[1.0, 0.9444444444444444, 1.0, 0.9444444444444444, 1.0, 1.0, 0.9444444444444444, 1.0, 0.6111111111111112, 1.0, 1.0, 1.0]
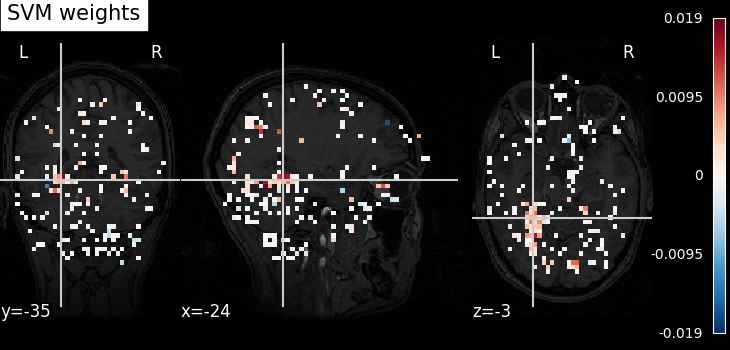
Visualize the results¶
Look at the SVC’s discriminating weights using
plot_stat_map
weight_img = decoder.coef_img_["face"]
from nilearn.plotting import plot_stat_map, show
plot_stat_map(weight_img, bg_img=haxby_dataset.anat[0], title="SVM weights")
show()
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_anova_svm.py:114: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Or we can plot the weights using view_img as a
dynamic html viewer
from nilearn.plotting import view_img
view_img(weight_img, bg_img=haxby_dataset.anat[0], title="SVM weights", dim=-1)
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.