Note
Go to the end to download the full example code or to run this example in your browser via Binder.
The haxby dataset: different multi-class strategies¶
We compare one vs all and one vs one multi-class strategies: the overall cross-validated accuracy and the confusion matrix.
Warning
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
import numpy as np
import pandas as pd
from nilearn import datasets
from nilearn.plotting import plot_matrix, show
Load the Haxby data dataset¶
# By default 2nd subject from haxby datasets will be fetched.
haxby_dataset = datasets.fetch_haxby()
# Print basic information on the dataset
print(f"Mask nifti images are located at: {haxby_dataset.mask}")
print(f"Functional nifti images are located at: {haxby_dataset.func[0]}")
func_filename = haxby_dataset.func[0]
mask_filename = haxby_dataset.mask
# Load the behavioral data that we will predict
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
y = labels["labels"]
run = labels["chunks"]
# Remove the rest condition, it is not very interesting
non_rest = y != "rest"
y = y[non_rest]
# Get the labels of the numerical conditions represented by the vector y
unique_conditions, order = np.unique(y, return_index=True)
# Sort the conditions by the order of appearance
unique_conditions = unique_conditions[np.argsort(order)]
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
Mask nifti images are located at: /home/runner/nilearn_data/haxby2001/mask.nii.gz
Functional nifti images are located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Prepare the fMRI data¶
from nilearn.maskers import NiftiMasker
# For decoding, standardizing is often very important
nifti_masker = NiftiMasker(
mask_img=mask_filename,
standardize="zscore_sample",
runs=run,
smoothing_fwhm=4,
memory="nilearn_cache",
memory_level=1,
verbose=1,
)
X = nifti_masker.fit_transform(func_filename)
# Remove the "rest" condition
X = X[non_rest]
run = run[non_rest]
[NiftiMasker.wrapped] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[NiftiMasker.wrapped] Loading data from
'/home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz'
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_multiclass.py:62: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[NiftiMasker.wrapped] Resampling mask
[NiftiMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.maskers.nifti_masker.filter_and_mask...
filter_and_mask('/home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz', <nibabel.nifti1.Nifti1Image object at 0x7f1eb0043a00>, { 'clean_args': None,
'clean_kwargs': {},
'cmap': 'gray',
'detrend': False,
'dtype': None,
'high_pass': None,
'high_variance_confounds': False,
'low_pass': None,
'reports': True,
'runs': 0 0
1 0
2 0
3 0
4 0
..
1447 11
1448 11
1449 11
1450 11
1451 11
Name: chunks, Length: 1452, dtype: int64,
'smoothing_fwhm': 4,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': None,
'target_affine': None,
'target_shape': None}, memory_level=1, memory=Memory(location=nilearn_cache/joblib), verbose=1, confounds=None, sample_mask=None, copy=True, dtype=None, sklearn_output_config=None)
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1eb0041780>
[NiftiMasker.wrapped] Smoothing images
[NiftiMasker.wrapped] Extracting region signals
[NiftiMasker.wrapped] Cleaning extracted signals
_________________________________________________filter_and_mask - 11.0s, 0.2min
Build the decoders, using scikit-learn¶
Here we use a Support Vector Classification, with a linear kernel, and a simple feature selection step
from sklearn.feature_selection import SelectKBest, f_classif
from sklearn.multiclass import OneVsOneClassifier, OneVsRestClassifier
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC
svc_ovo = OneVsOneClassifier(
Pipeline(
[
("anova", SelectKBest(f_classif, k=500)),
("svc", SVC(kernel="linear")),
]
)
)
svc_ova = OneVsRestClassifier(
Pipeline(
[
("anova", SelectKBest(f_classif, k=500)),
("svc", SVC(kernel="linear")),
]
)
)
Now we compute cross-validation scores¶
from sklearn.model_selection import cross_val_score
cv_scores_ovo = cross_val_score(svc_ovo, X, y, cv=5, verbose=1)
cv_scores_ova = cross_val_score(svc_ova, X, y, cv=5, verbose=1)
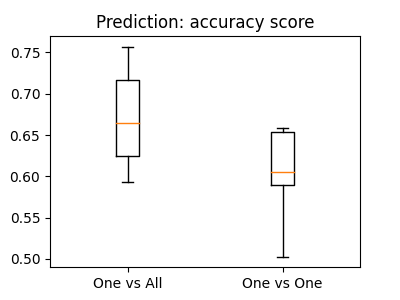
print("OvO:", cv_scores_ovo.mean())
print("OvA:", cv_scores_ova.mean())
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 9.0s finished
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 4.9s finished
OvO: 0.601855088049469
OvA: 0.6712058072321548
Plot barplots of the prediction scores¶
from matplotlib import pyplot as plt
plt.figure(figsize=(4, 3))
plt.boxplot([cv_scores_ova, cv_scores_ovo])
plt.xticks([1, 2], ["One vs All", "One vs One"])
plt.title("Prediction: accuracy score")
Text(0.5, 1.0, 'Prediction: accuracy score')
Plot a confusion matrix¶
We fit on the first 10 runs and plot a confusion matrix on the last 2 runs
from sklearn.metrics import confusion_matrix
svc_ovo.fit(X[run < 10], y[run < 10])
y_pred_ovo = svc_ovo.predict(X[run >= 10])
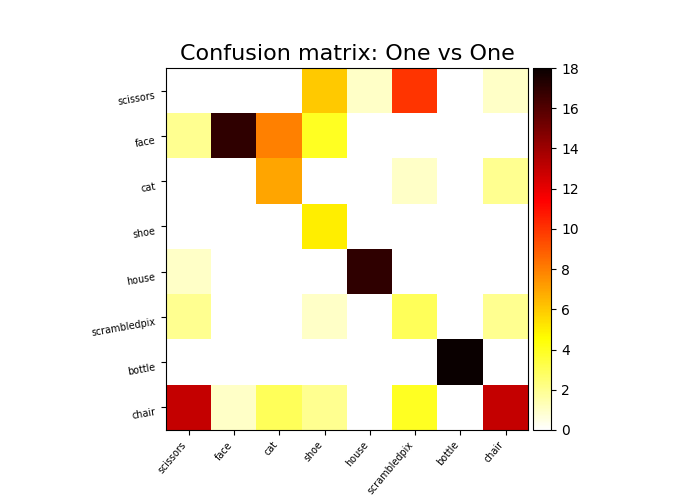
plot_matrix(
confusion_matrix(y_pred_ovo, y[run >= 10]),
labels=unique_conditions,
title="Confusion matrix: One vs One",
cmap="inferno",
)
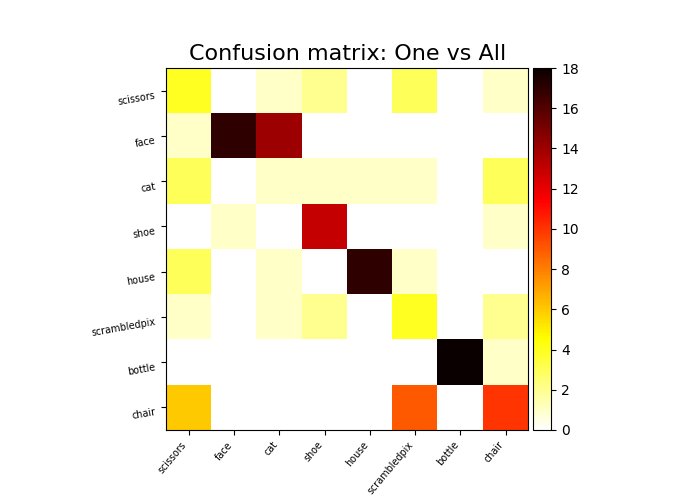
svc_ova.fit(X[run < 10], y[run < 10])
y_pred_ova = svc_ova.predict(X[run >= 10])
plot_matrix(
confusion_matrix(y_pred_ova, y[run >= 10]),
labels=unique_conditions,
title="Confusion matrix: One vs All",
cmap="inferno",
)
show()
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_multiclass.py:146: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (0 minutes 31.914 seconds)
Estimated memory usage: 2474 MB