Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Generate an events.tsv file for the NeuroSpin localizer task¶
Create a BIDS-compatible events.tsv file from onset/trial-type information.
The protocol described is the so-called “ARCHI Standard” functional localizer task.
For details on the task, please see Pinel et al.[1].
Define the onset times in seconds. These are typically extracted from the stimulation software used, but we will use hardcoded values in this example.
# fmt: off
onsets = [
0.0, 2.4, 8.7, 11.4, 15.0, 18.0, 20.7, 23.7, 26.7, 29.7,
33.0, 35.4, 39.0, 41.7, 44.7, 48.0, 56.4, 59.7, 62.4, 69.0,
71.4, 75.0, 83.4, 87.0, 89.7, 96.0, 108.0, 116.7, 119.4, 122.7,
125.4, 131.4, 135.0, 137.7, 140.4, 143.4, 146.7, 149.4, 153.0, 156.0,
159.0, 162.0, 164.4, 167.7, 170.4, 173.7, 176.7, 188.4, 191.7, 195.0,
198.0, 201.0, 203.7, 207.0, 210.0, 212.7, 215.7, 218.7, 221.4, 224.7,
227.7, 230.7, 234.0, 236.7, 246.0, 248.4, 251.7, 254.7, 257.4, 260.4,
264.0, 266.7, 269.7, 275.4, 278.4, 284.4, 288.0, 291.0, 293.4, 296.7,
]
# fmt: on
Associated trial types: these are numbered between 0 and 9, hence corresponding to 10 different conditions.
# fmt: off
trial_type_idx = [
7, 7, 0, 2, 9, 4, 9, 3, 5, 9, 1, 6, 8, 8, 6, 6, 8, 0, 3, 4, 5, 8, 6,
2, 9, 1, 6, 5, 9, 1, 7, 8, 6, 6, 1, 2, 9, 0, 7, 1, 8, 2, 7, 8, 3, 6,
0, 0, 6, 8, 7, 7, 1, 1, 1, 5, 5, 0, 7, 0, 4, 2, 7, 9, 8, 0, 6, 3, 3,
7, 1, 0, 0, 4, 1, 9, 8, 4, 9, 9
]
# fmt: on
We may want to map these indices to explicit condition names. For that, we define a list of 10 strings.
condition_ids = [
"horizontal checkerboard",
"vertical checkerboard",
"right button press, auditory instructions",
"left button press, auditory instructions",
"right button press, visual instructions",
"left button press, visual instructions",
"mental computation, auditory instructions",
"mental computation, visual instructions",
"visual sentence",
"auditory sentence",
]
trial_types = [condition_ids[i] for i in trial_type_idx]
We must also define a duration (required by BIDS conventions). In this case, all trials lasted one second, except for button response that are modeled as events with instantaneous duration (0 second).
null_duration_trials = [2, 3, 4, 5]
durations = []
for i in trial_type_idx:
if i in null_duration_trials:
durations.append(0)
else:
durations.append(1)
Form a pandas DataFrame from this information.
import pandas as pd
events = pd.DataFrame(
{
"trial_type": trial_types,
"onset": onsets,
"duration": durations,
}
)
Take a look at the new DataFrame
Export them to a tsv file.
from pathlib import Path
output_dir = Path.cwd() / "results" / "plot_write_events_file"
output_dir.mkdir(exist_ok=True, parents=True)
tsvfile = output_dir / "localizer_events.tsv"
events.to_csv(tsvfile, sep="\t", index=False)
print(f"The event information has been saved to {tsvfile}")
The event information has been saved to /home/runner/work/nilearn/nilearn/examples/04_glm_first_level/results/plot_write_events_file/localizer_events.tsv
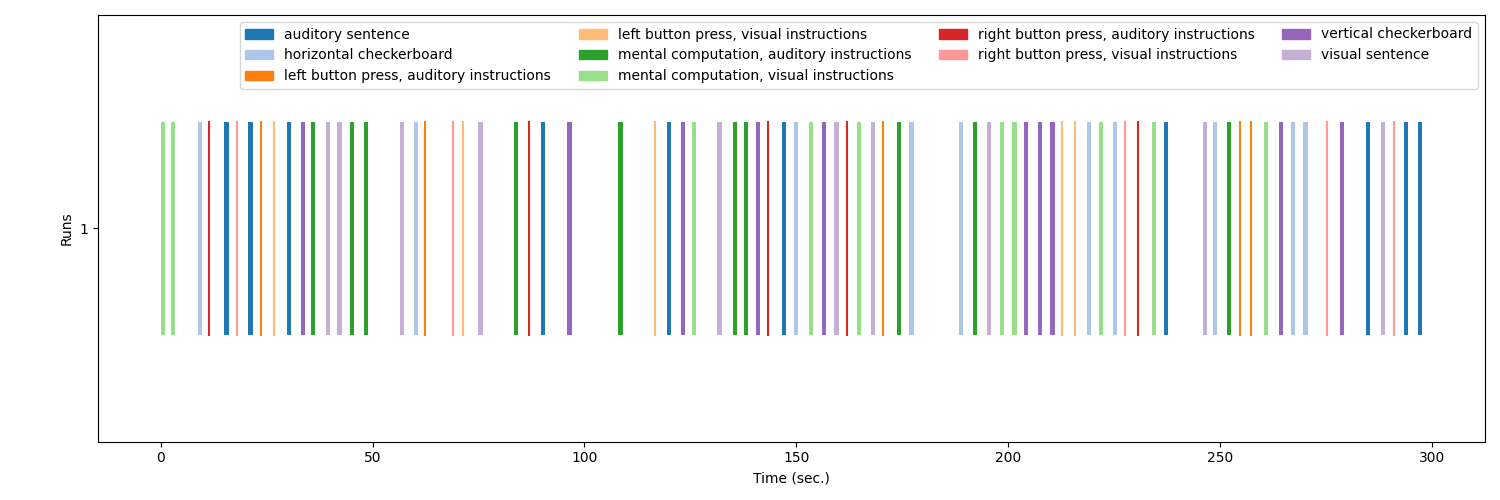
Optionally, the events can be visualized using the
plot_event function.
from nilearn.plotting import plot_event, show
fig = plot_event(events, figsize=(15, 5))
fig.suptitle("Events")
show()
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_write_events_file.py:107: UserWarning:
The following conditions contain events with null duration:
- 'left button press, auditory instructions'
- 'left button press, visual instructions'
- 'right button press, auditory instructions'
- 'right button press, visual instructions'
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_write_events_file.py:109: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Parametric modulation¶
We may want to modulate the way we model our events in our fMRI analysis. This type of parametric modulation can be done by adding a “modulation” column to the dataframe containing our events.
Here we will assume that when a trial is the same condition as the previous one, it will elicit a less intense response.
modulations = []
conditions_to_modulate = [
"horizontal checkerboard",
"vertical checkerboard",
"mental computation, auditory instructions",
"mental computation, visual instructions",
"visual sentence",
"auditory sentence",
]
for i, trial in enumerate(trial_types):
if (
i > 0
and trial in conditions_to_modulate
and trial == trial_types[i - 1]
):
modulations.append(0.5)
else:
modulations.append(1)
modulated_events = pd.DataFrame(
{
"trial_type": trial_types,
"onset": onsets,
"duration": durations,
"modulation": modulations,
}
)
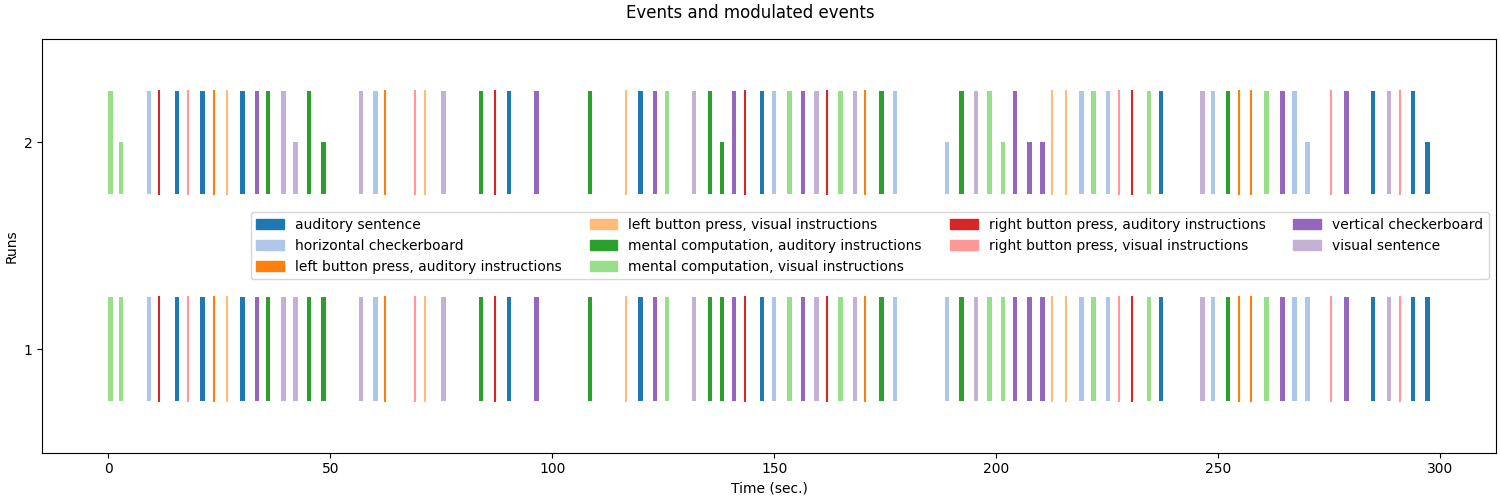
# Now lets plot the modulated and unmodulated events.
fig = plot_event([events, modulated_events], figsize=(15, 5))
fig.suptitle("Events and modulated events")
show()
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_write_events_file.py:151: UserWarning:
The following conditions contain events with null duration:
- 'left button press, auditory instructions'
- 'left button press, visual instructions'
- 'right button press, auditory instructions'
- 'right button press, visual instructions'
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_write_events_file.py:151: UserWarning:
The following conditions contain events with null duration:
- 'left button press, auditory instructions'
- 'left button press, visual instructions'
- 'right button press, auditory instructions'
- 'right button press, visual instructions'
[plot_event] A 'modulation' column was found in the given events data and is
used.
/home/runner/work/nilearn/nilearn/examples/04_glm_first_level/plot_write_events_file.py:153: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Note
See how the modulation affects the height of the events.
References¶
Total running time of the script: (0 minutes 4.440 seconds)
Estimated memory usage: 107 MB