Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Plotting tools in nilearn¶
Nilearn comes with a set of plotting functions for easy visualization of Nifti-like images such as statistical maps mapped onto anatomical images or onto glass brain representation, anatomical images, functional/EPI images, region specific mask images.
See Plotting brain images for more details.
We will first retrieve data from nilearn provided (general-purpose) datasets.
from nilearn import datasets
# haxby dataset to have EPI images and masks
haxby_dataset = datasets.fetch_haxby()
# print basic information on the dataset
print(
f"First subject anatomical nifti image (3D) is at: {haxby_dataset.anat[0]}"
)
print(
f"First subject functional nifti image (4D) is at: {haxby_dataset.func[0]}"
)
haxby_anat_filename = haxby_dataset.anat[0]
haxby_mask_filename = haxby_dataset.mask_vt[0]
haxby_func_filename = haxby_dataset.func[0]
# one motor activation map
stat_img = datasets.load_sample_motor_activation_image()
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
First subject anatomical nifti image (3D) is at: /home/runner/nilearn_data/haxby2001/subj2/anat.nii.gz
First subject functional nifti image (4D) is at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Nilearn plotting functions¶
Plotting statistical maps: plot_stat_map¶
from nilearn import plotting
Visualizing t-map image on EPI template with manual positioning of coordinates using cut_coords given as a list
plotting.plot_stat_map(
stat_img, threshold=3, title="plot_stat_map", cut_coords=[36, -27, 66]
)
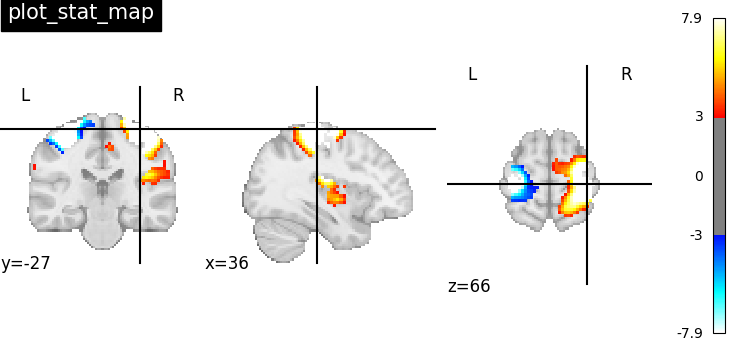
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee8a542b0>
It’s also possible to visualize volumes in a LR-flipped “radiological” view Just set radiological=True
plotting.plot_stat_map(
stat_img,
threshold=3,
title="plot_stat_map",
cut_coords=[36, -27, 66],
radiological=True,
)
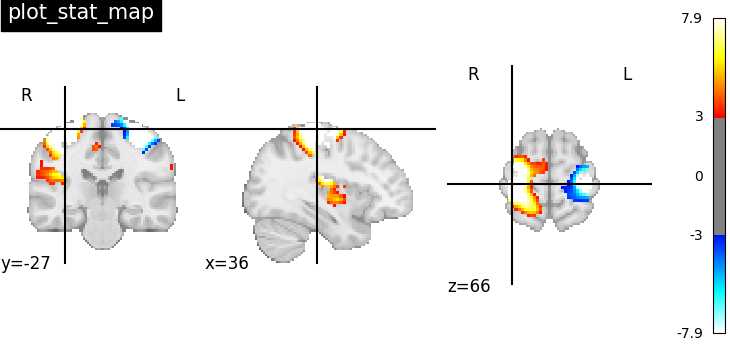
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ef14535b0>
Making interactive visualizations: view_img¶
An alternative to plot_stat_map is to use
view_img that gives more interactive
visualizations in a web browser. See Interactive visualization of statistical map slices
for more details.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.
uncomment this to open the plot in a web browser: view.open_in_browser()
It’s also possible to visualize volumes in a LR-flipped “radiological” view Just set radiological=True
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/numpy/_core/fromnumeric.py:868: UserWarning:
Warning: 'partition' will ignore the 'mask' of the MaskedArray.
uncomment this to open the plot in a web browser: view_radio.open_in_browser()
Plotting statistical maps in a glass brain: plot_glass_brain¶
Now, the t-map image is mapped on glass brain representation where glass brain is always a fixed background template
plotting.plot_glass_brain(stat_img, title="plot_glass_brain", threshold=3)
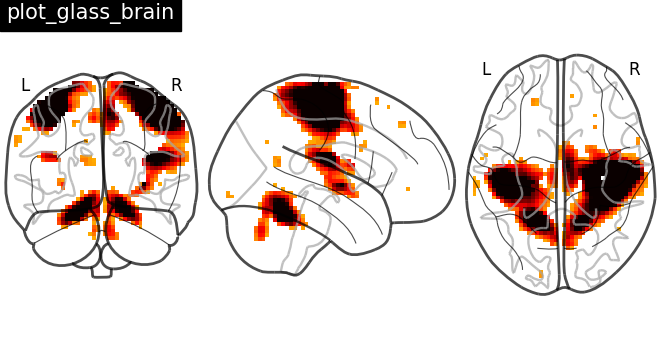
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f1ed010e530>
Plotting anatomical images: plot_anat¶
Visualizing anatomical image of haxby dataset
plotting.plot_anat(haxby_anat_filename, title="plot_anat")
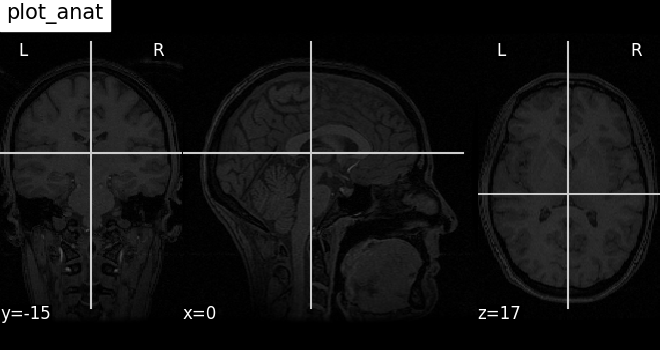
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0af8c0a0>
Plotting ROIs (here the mask): plot_roi¶
Visualizing ventral temporal region image from haxby dataset overlaid on subject specific anatomical image with coordinates positioned automatically on region of interest (roi)
plotting.plot_roi(
haxby_mask_filename, bg_img=haxby_anat_filename, title="plot_roi"
)
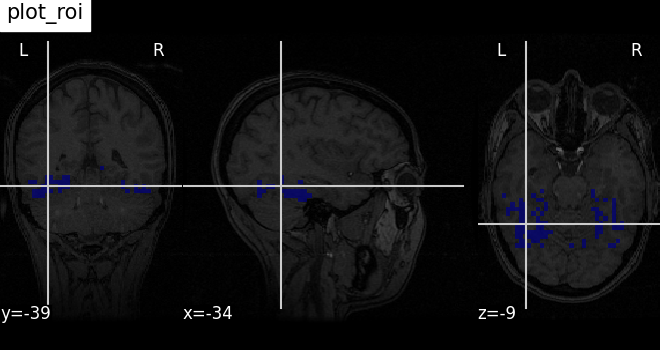
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0c7d03d0>
Plotting EPI image: plot_epi¶
# Import image processing tool
from nilearn import image
# Compute the voxel_wise mean of functional images across time.
# Basically reducing the functional image from 4D to 3D
mean_haxby_img = image.mean_img(haxby_func_filename)
# Visualizing mean image (3D)
plotting.plot_epi(mean_haxby_img, title="plot_epi")
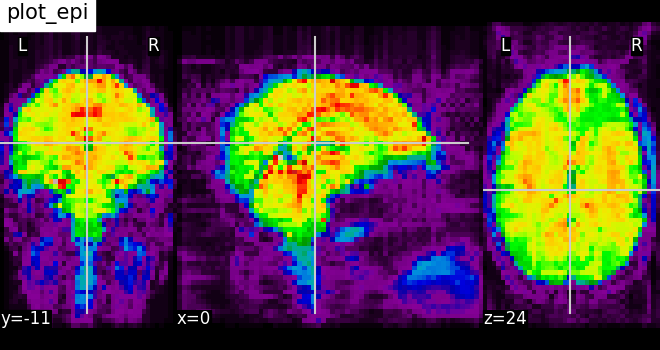
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee8e3a140>
A call to plotting.show is needed to display the plots when running in script mode (ie outside IPython)
/home/runner/work/nilearn/nilearn/examples/01_plotting/plot_demo_plotting.py:140: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Thresholding plots¶
Using threshold value alongside with vmin and vmax parameters
enable us to mask certain values in the image.
Plotting without threshold¶
plotting.plot_stat_map(
stat_img,
display_mode="ortho",
cut_coords=[36, -27, 60],
title="No plotting threshold",
)
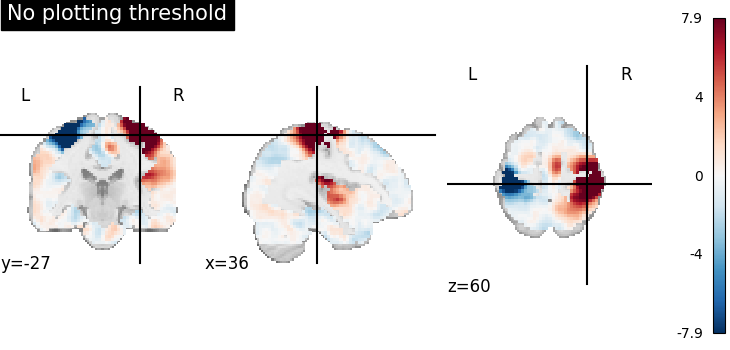
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0c2f48e0>
Plotting threshold set to 1¶
When plotting threshold is set to 1, the values between -1 and 1 are masked in the plot.
plotting.plot_stat_map(
stat_img,
threshold=1,
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1",
)
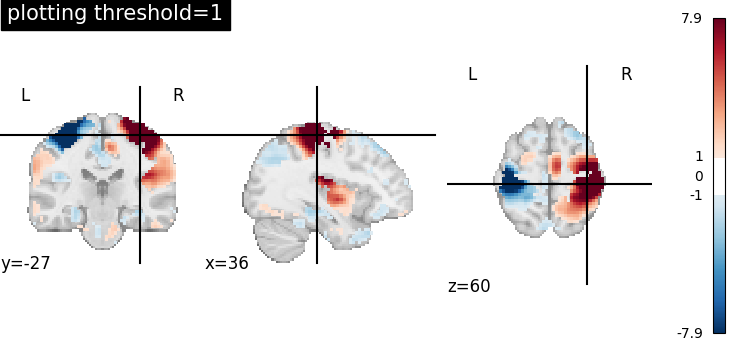
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ef1474850>
Plotting threshold set to 1 with vmin=0¶
Setting vmin=0, it is possible to plot only positive image values.
plotting.plot_stat_map(
stat_img,
threshold=1,
cmap="inferno",
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1, vmin=0",
vmin=0,
)
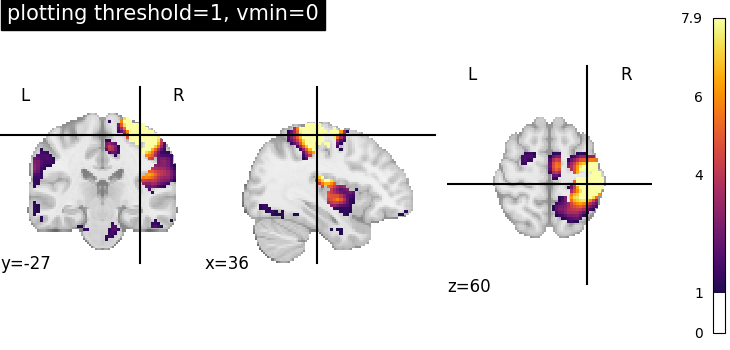
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee8b11f30>
Plotting threshold set to 1 with vmax=0¶
Setting vmax=0, it is possible to plot only negative image values.
plotting.plot_stat_map(
stat_img,
threshold=1,
cmap="inferno",
display_mode="ortho",
cut_coords=[36, -27, 60],
title="plotting threshold=1, vmax=0",
vmax=0,
)
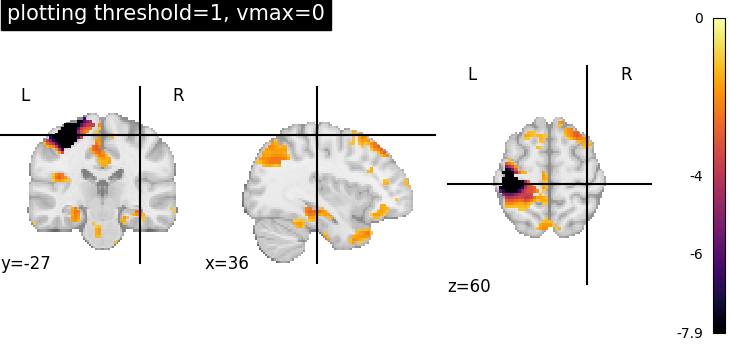
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0c7ef6a0>
Visualizing without a colorbar on the right side¶
The argument colorbar should be set to False to show plots without
a colorbar on the right side.
plotting.plot_stat_map(
stat_img,
display_mode="ortho",
cut_coords=[36, -27, 60],
colorbar=False,
title="no colorbar",
)
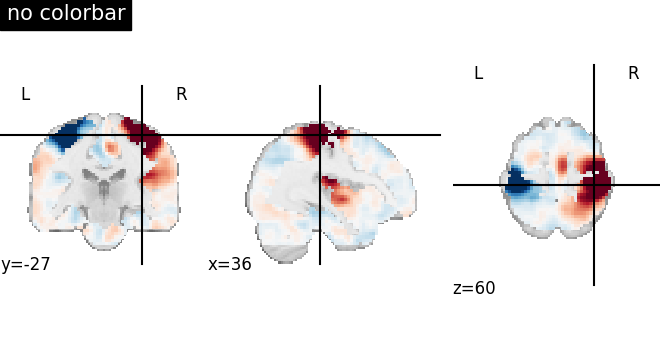
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0c2bce80>
Total running time of the script: (0 minutes 22.675 seconds)
Estimated memory usage: 1015 MB