Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Glass brain plotting in nilearn (all options)¶
The first part of this example goes through different options of the
plot_glass_brain function (including plotting
negative values).
The second part goes through same options but selected of the same glass brain function but plotting is seen with contours.
See Plotting brain images for more plotting functionalities and Section 4.3 for more details about display objects in Nilearn.
Also, see load_sample_motor_activation_image
for details about the plotting data and associated meta-data.
Load the data¶
We will use a motor activation contrast map distributed with Nilearn.
from nilearn import datasets
stat_img = datasets.load_sample_motor_activation_image()
# stat_img is just the name of the image file
stat_img
'/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/nilearn/datasets/data/image_10426.nii.gz'
Demo glass brain plotting¶
By default, plot_glass_brain uses a display mode
called ‘ortho’ which results in three projections. It is equivalent to
specify display_mode='ortho' in
plot_glass_brain. Note that depending on the
value of display_mode, different display objects are returned. Here,
a OrthoProjector is returned.
from nilearn.plotting import plot_glass_brain, show
# Whole brain sagittal cuts and map is thresholded at 3
plot_glass_brain(stat_img, threshold=3)
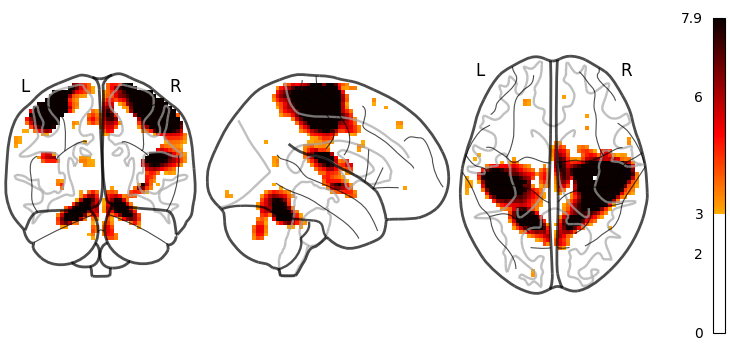
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f1ee8466b00>
The same figure, without a colorbar,
can be produced by setting colorbar=False.
plot_glass_brain(stat_img, threshold=3, colorbar=False)
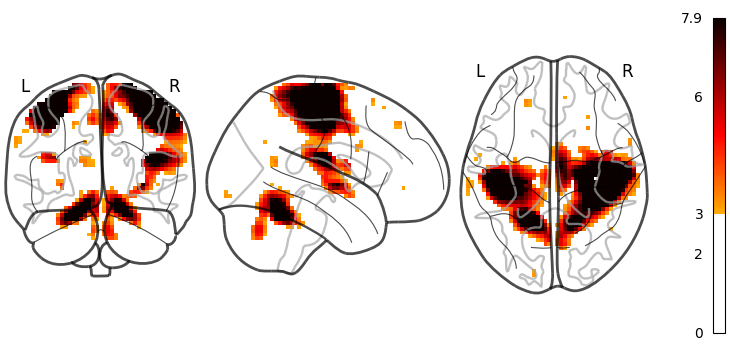
<nilearn.plotting.displays._projectors.OrthoProjector object at 0x7f1f0ca6f280>
Here, we show how to set a black background, and we only view sagittal and
axial projections by setting display_mode='xz', which returns a
XZProjector.
plot_glass_brain(
stat_img,
title="plot_glass_brain",
black_bg=True,
display_mode="xz",
threshold=3,
)
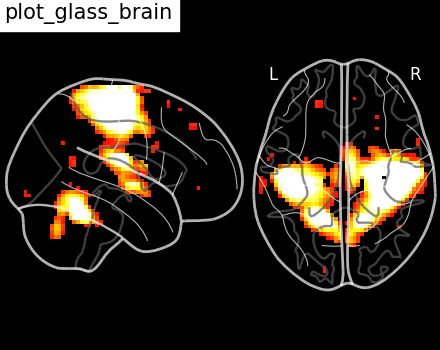
<nilearn.plotting.displays._projectors.XZProjector object at 0x7f1f0ce111e0>
We can also plot the sign of the activation by setting plot_abs=False.
Additionally, we only visualize coronal and axial projections by setting
display_mode='yz' which returns a
YZProjector.
plot_glass_brain(stat_img, threshold=0, plot_abs=False, display_mode="yz")
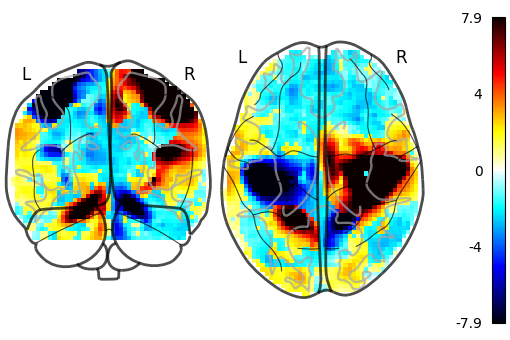
<nilearn.plotting.displays._projectors.YZProjector object at 0x7f1ee8879480>
Setting plot_abs=True and display_mode='yx' (returns a
YXProjector).
plot_glass_brain(stat_img, threshold=3, plot_abs=True, display_mode="yx")
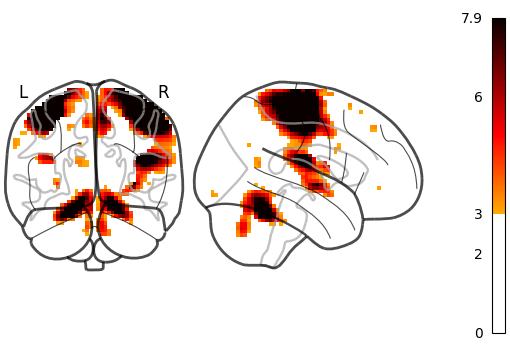
<nilearn.plotting.displays._projectors.YXProjector object at 0x7f1ee8ba42b0>
We can control the limits of the colormap and colorbar by setting vmin
and vmax.
Note that we use a non-diverging colormap here
since the colorbar will not be centered around zero.
# only plot positive values
plot_glass_brain(
stat_img,
plot_abs=False,
display_mode="yz",
vmin=0,
threshold=2,
symmetric_cbar=False,
cmap="inferno",
title="only plot positive values",
)
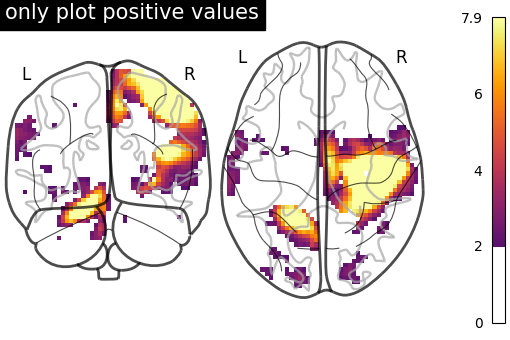
<nilearn.plotting.displays._projectors.YZProjector object at 0x7f1f0c395b40>
Here we set vmin to the threshold to use the full color range instead of
losing colors due to the thresholding.
plot_glass_brain(
stat_img,
plot_abs=False,
display_mode="yz",
vmin=2,
threshold=2,
symmetric_cbar=False,
cmap="inferno",
title="vmin == threshold",
)
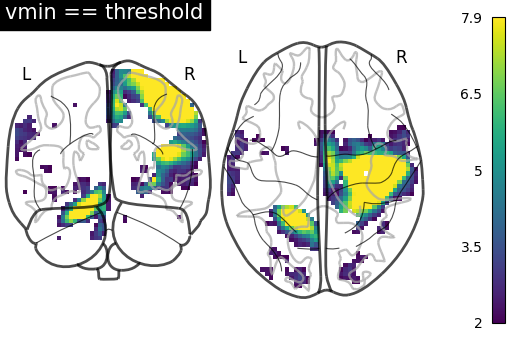
<nilearn.plotting.displays._projectors.YZProjector object at 0x7f1ee8b5d930>
Different projections for the left and right hemispheres¶
In the previous section we saw a few projection modes, which are controlled
by setting the argument display_mode of
plot_glass_brain. In this section, we will show
some additional possibilities. For example, setting display_mode='lzr'
enables an hemispheric sagittal view. The display object returned is then a
LZRProjector.
plot_glass_brain(
stat_img,
title='plot_glass_brain with display_mode="lzr"',
black_bg=True,
display_mode="lzr",
threshold=3,
)
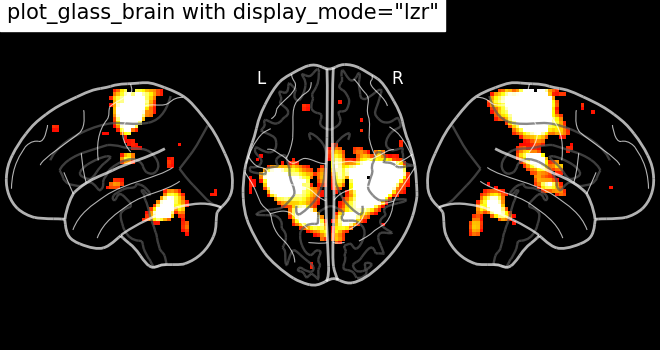
<nilearn.plotting.displays._projectors.LZRProjector object at 0x7f1f0c3e9000>
display_mode='lyrz' returns a
LYRZProjector object.
plot_glass_brain(
stat_img,
threshold=0,
title='plot_glass_brain with display_mode="lyrz"',
plot_abs=False,
display_mode="lyrz",
)
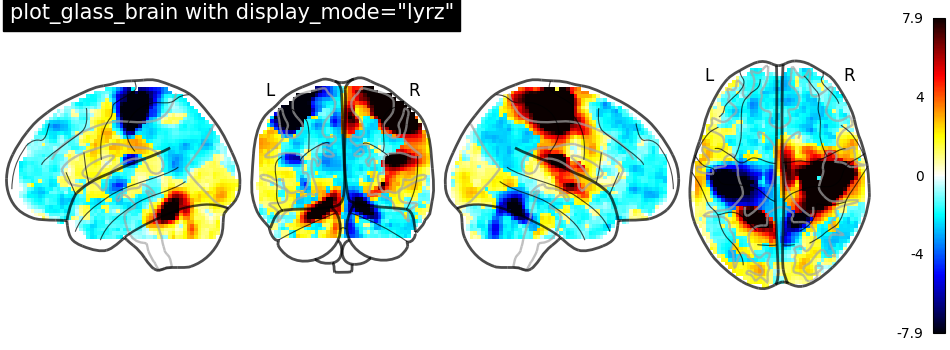
<nilearn.plotting.displays._projectors.LYRZProjector object at 0x7f1ee8abb490>
If you are only interested in single projections, you can set
display_mode to
‘x’ (returns a XProjector),
‘y’ (returns a YProjector),
‘z’ (returns a ZProjector),
‘l’ (returns a LProjector), or
‘r’ (returns a RProjector).
plot_glass_brain(
stat_img,
threshold=0,
title='display_mode="x"',
plot_abs=True,
display_mode="x",
)
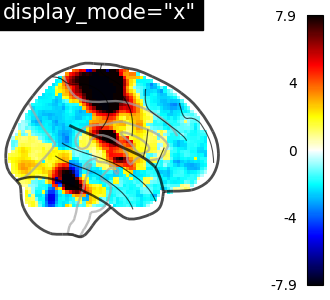
<nilearn.plotting.displays._projectors.XProjector object at 0x7f1f0cdf9fc0>
Contours and with fillings¶
The display objects returned by plot_glass_brain
all inherit from the OrthoProjector
and enable further customization of the figures.
In this example, we focus on using methods
add_contours and
title. First, we
save the display object (here a
LZRYProjector) into a variable named
display. Note that we set the first argument to None since we
want an empty glass brain to plot the statistical maps with
add_contours.
display = plot_glass_brain(None, display_mode="lzry")
# Here, we project statistical maps
display.add_contours(stat_img)
# and add a title
display.title('"stat_img" on glass brain without threshold')
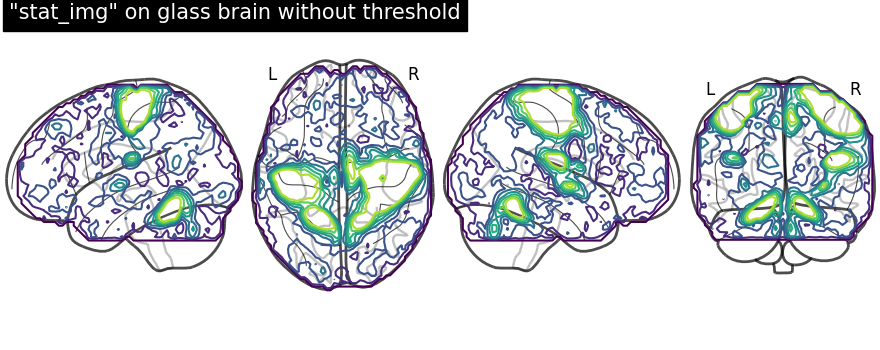
We can fill the contours by setting filled=True. Note that we are not
specifying levels here
display = plot_glass_brain(None, display_mode="lzry")
# Here, we project statistical maps with filled=True
display.add_contours(stat_img, filled=True)
# and add a title
display.title("Same map but with fillings in the contours")
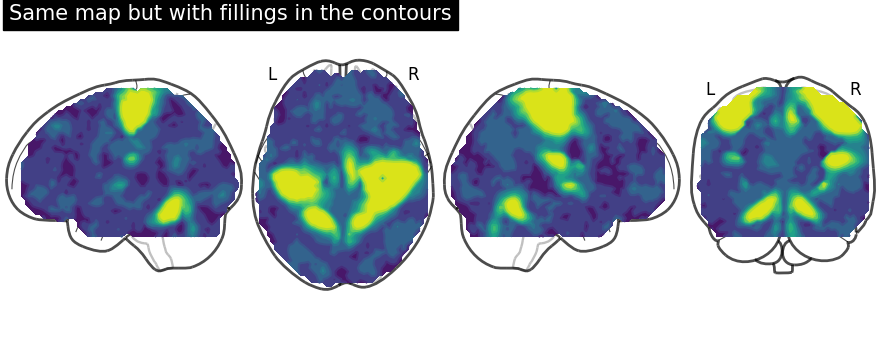
Here, we input a specific level (cut-off) in the statistical map. In other words, we are thresholding our statistical map.
We set the threshold using a parameter of method
add_contours called
levels which value is given as a list and we choose the color to be red.
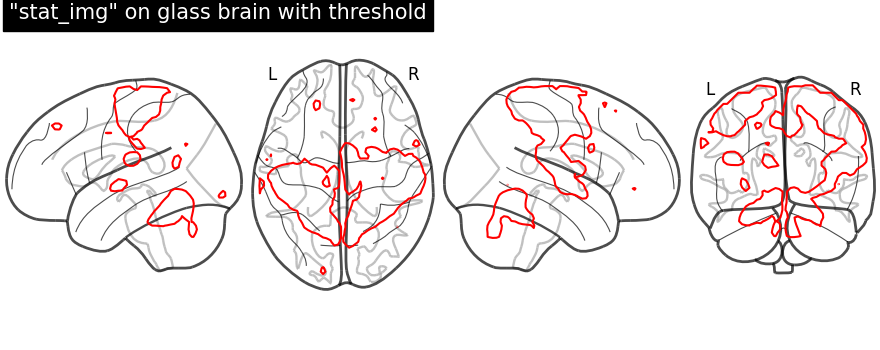
display = plot_glass_brain(None, display_mode="lzry")
display.add_contours(stat_img, levels=[3.0], colors="r")
display.title('"stat_img" on glass brain with threshold')
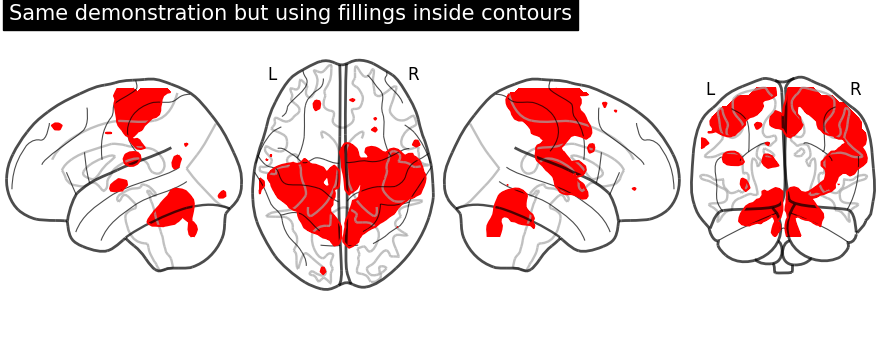
Plotting with same demonstration but fill the contours (by setting
filled=True).
display = plot_glass_brain(None, display_mode="lzry")
display.add_contours(stat_img, filled=True, levels=[3.0], colors="r")
display.title("Same demonstration but using fillings inside contours")
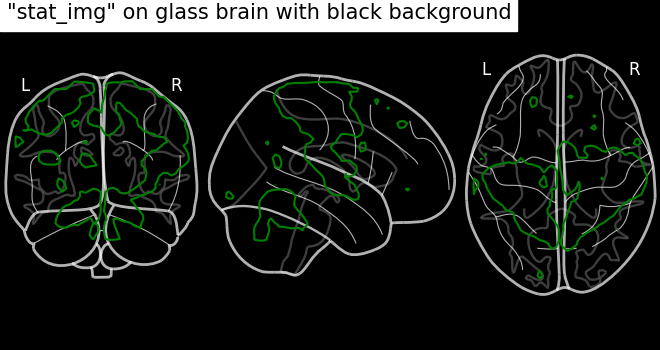
Plotting with black background, black_bg should be set to True
through plot_glass_brain.
# We can set black background using black_bg=True
display = plot_glass_brain(None, black_bg=True)
display.add_contours(stat_img, levels=[3.0], colors="g")
display.title('"stat_img" on glass brain with black background')
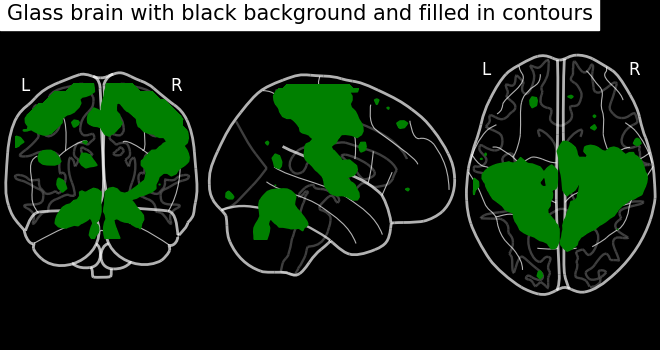
Black background plotting with filled in contours.
display = plot_glass_brain(None, black_bg=True)
display.add_contours(stat_img, filled=True, levels=[3.0], colors="g")
display.title("Glass brain with black background and filled in contours")
Contour projections in both hemispheres¶
The key argument to vary here is display_mode for hemispheric plotting.
Here, we set display_mode='lr' for both hemispheric plots. Note that a
LRProjector is returned.
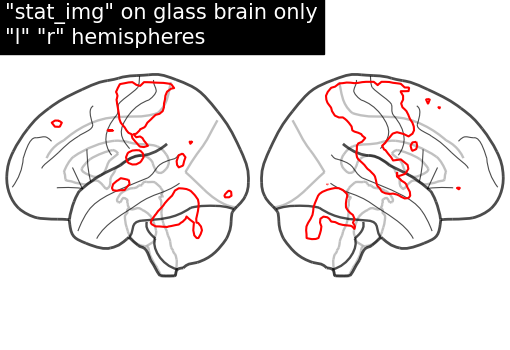
display = plot_glass_brain(None, display_mode="lr")
display.add_contours(stat_img, levels=[3.0], colors="r")
display.title('"stat_img" on glass brain only\n"l" "r" hemispheres')
Filled contours in both hemispheric plotting, by adding filled=True.
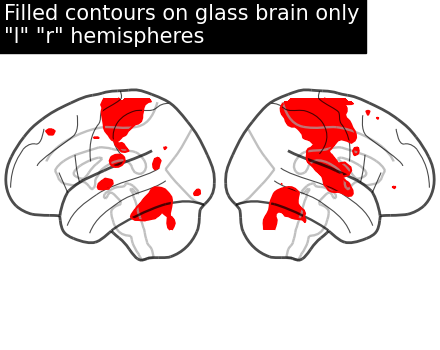
display = plot_glass_brain(None, display_mode="lr")
display.add_contours(stat_img, filled=True, levels=[3.0], colors="r")
display.title('Filled contours on glass brain only\n"l" "r" hemispheres')
With positive and negative signs of activations with plot_abs in
plot_glass_brain.
By default parameter plot_abs is True and sign of activations
can be displayed by changing plot_abs to False. Note that we also
specify display_mode='lyr' which returns a
LYRProjector display object.
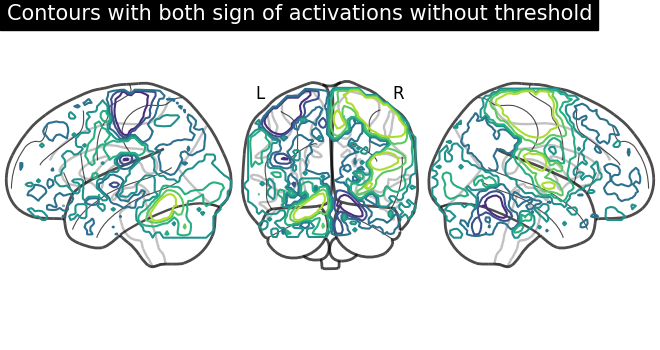
display = plot_glass_brain(None, plot_abs=False, display_mode="lyr")
display.add_contours(stat_img)
display.title("Contours with both sign of activations without threshold")
Now, adding filled=True to get positive and negative sign activations
with fillings in the contours.
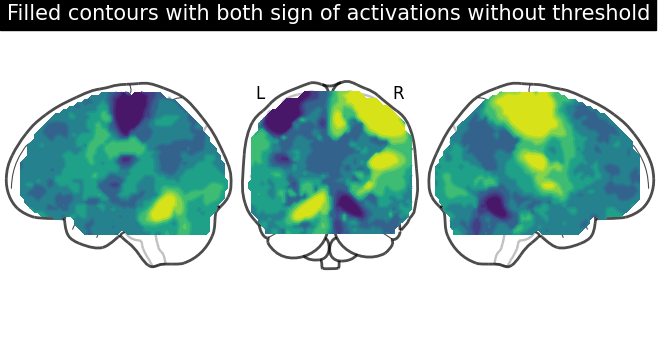
display = plot_glass_brain(None, plot_abs=False, display_mode="lyr")
display.add_contours(stat_img, filled=True)
display.title(
"Filled contours with both sign of activations without threshold"
)
Displaying both signs (positive and negative) of activations with threshold
meaning thresholding by adding an argument levels in method
add_contours.
We give two values through the argument levels which corresponds to the
thresholds of the contour we want to draw: One is positive and the other one
is negative. We give a list of colors as argument to associate a
different color to each contour. Additionally, we also choose to plot
contours with thick line widths. For linewidths, one value would be
enough so that same value is used for both contours.
import numpy as np
display = plot_glass_brain(None, plot_abs=False, display_mode="lzry")
display.add_contours(
stat_img, levels=[-2.8, 3.0], colors=["b", "r"], linewidths=4.0
)
display.title("Contours with sign of activations with threshold")
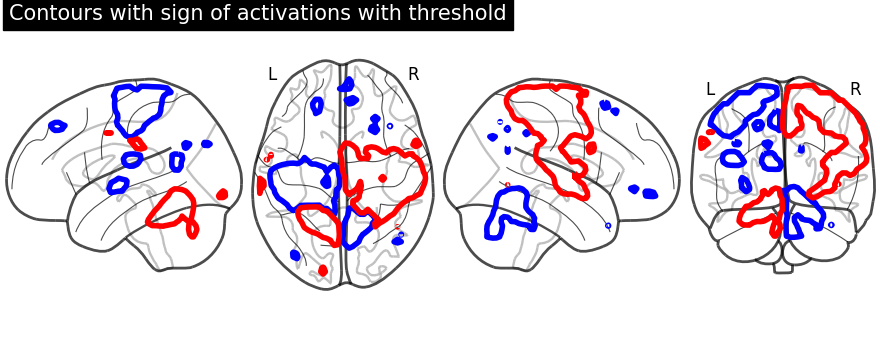
Same display demonstration as above but adding filled=True to get
fillings inside the contours.
Unlike in previous plot, here we specify each sign at a time. We call negative values display first followed by positive values display.
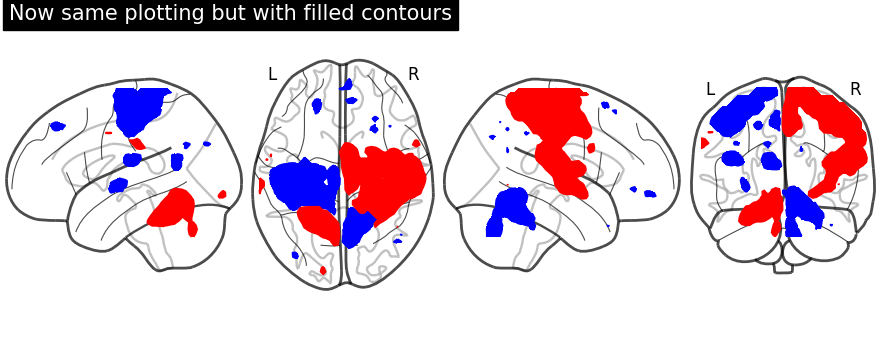
First, we fetch our display object with same parameters used as above. Then, we plot negative sign of activation with levels given as negative activation value in a list. Upper bound should be kept to -infinity. Next, using the same display object, we plot positive sign of activation.
display = plot_glass_brain(None, plot_abs=False, display_mode="lzry")
display.add_contours(stat_img, filled=True, levels=[-np.inf, -2.8], colors="b")
display.add_contours(stat_img, filled=True, levels=[3.0], colors="r")
display.title("Now same plotting but with filled contours")
# Finally, displaying them
show()
/home/runner/work/nilearn/nilearn/examples/01_plotting/plot_demo_glass_brain_extensive.py:328: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (0 minutes 26.711 seconds)
Estimated memory usage: 112 MB