Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.image.mean_img¶
- nilearn.image.mean_img(imgs, target_affine=None, target_shape=None, verbose=0, n_jobs=1, copy_header=True)[source]¶
Compute the mean over images.
This can be a mean over time or the 4th dimension for a volume, or the 2nd dimension for a surface image.
Note that if list of 4D volume images (or 2D surface images) are given, the mean of each image is computed separately, and the resulting mean is computed after.
- Parameters:
- imgsNiimg-like or
SurfaceImageobject, or iterable of Niimg-like orSurfaceImage. Images to be averaged over ‘time’ (see Input and output: neuroimaging data representation for a detailed description of the valid input types).
- target_affine3x3 or a 4x4 array-like, or None, default=None
If specified, the image is resampled corresponding to this new affine. Ignored for
SurfaceImage.- target_shape
tupleorlistor None, default=None If specified, the image will be resized to match this new shape. len(target_shape) must be equal to 3.
Note
If target_shape is specified, a target_affine of shape (4, 4) must also be given.
Ignored for
SurfaceImage.- verbose
boolorint, default=0 Verbosity level (
0orFalsemeans no message).- n_jobs
int, default=1 The number of CPUs to use to do the computation (-1 means ‘all CPUs’). Ignored for
SurfaceImage.- copy_header
bool, default=True Indicated if the header of the reference image should be used to create the new image. Ignored for
SurfaceImage.Added in Nilearn 0.11.0.
- imgsNiimg-like or
- Returns:
Nifti1ImageorSurfaceImageMean image.
See also
nilearn.image.math_imgFor more general operations on images.
Examples using nilearn.image.mean_img¶
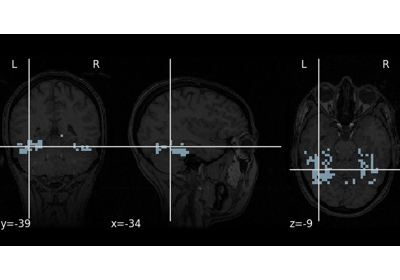
Intro to GLM Analysis: a single-run, single-subject fMRI dataset
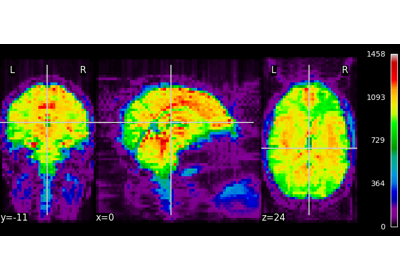
Decoding of a dataset after GLM fit for signal extraction
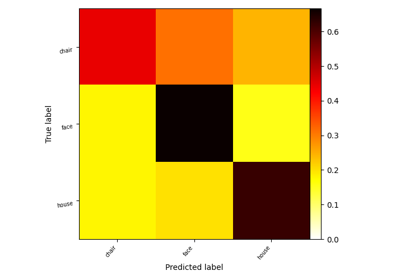
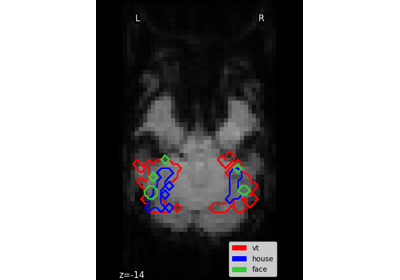
Decoding with FREM: face vs house vs chair object recognition
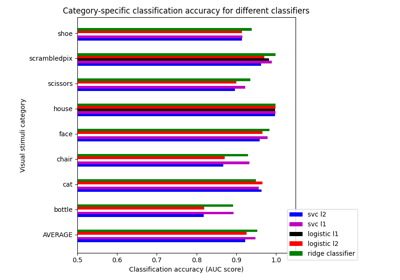
Different classifiers in decoding the Haxby dataset
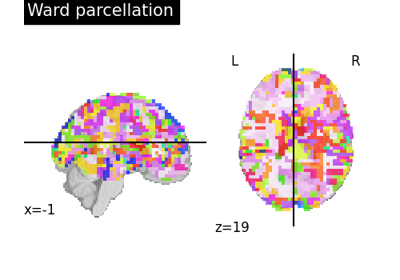
Clustering methods to learn a brain parcellation from fMRI
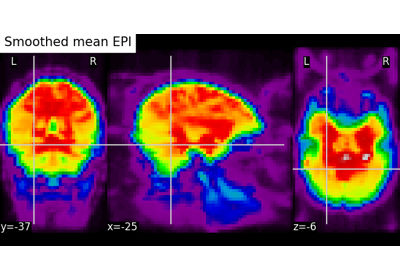
Computing a Region of Interest (ROI) mask manually
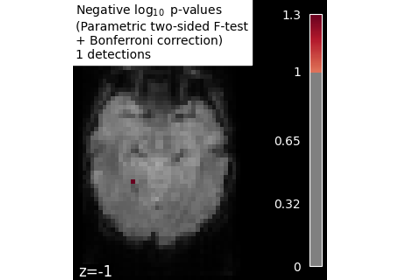
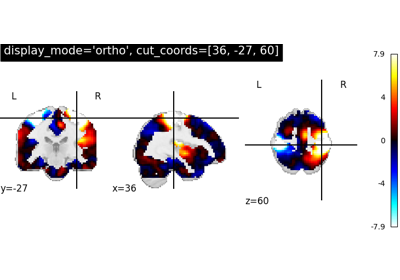
Massively univariate analysis of face vs house recognition
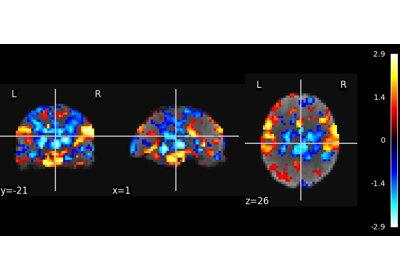
Multivariate decompositions: Independent component analysis of fMRI