Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Understanding Decoder¶
Nilearn’s Decoder object is a composite estimator
that does several things under the hood and can hence be a bit difficult to
understand at first.
This example aims to provide a clear understanding of the
Decoder object by demonstrating these steps via a
Scikit-Learn pipeline.
We will use the Haxby et al.[1] dataset where the participants were shown images of 8 different types as described in the Decoding with ANOVA + SVM: face vs house in the Haxby dataset example. We will train a classifier to predict the label of the object in the stimulus image based on the subject’s fMRI data from the Ventral Temporal cortex.
Load the Haxby dataset¶
from nilearn import datasets
# By default 2nd subject data will be fetched on which we run our analysis
haxby_dataset = datasets.fetch_haxby()
fmri_img = haxby_dataset.func[0]
# Pick the mask that we will use to extract the data from Ventral Temporal
# cortex
mask_vt = haxby_dataset.mask_vt[0]
# Load the behavioral data
import pandas as pd
from nilearn.image import index_img
behavioral_data = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
labels = behavioral_data["labels"]
# Keep the trials corresponding to all the labels except the ``rest`` ones
labels_mask = labels != "rest"
y = labels[labels_mask]
y = y.to_numpy()
# Load run information
run = behavioral_data["chunks"][labels_mask]
run = run.to_numpy()
# Also keep the fmri data corresponding to these labels
fmri_img = index_img(fmri_img, labels_mask)
# Overview of the input data
import numpy as np
n_labels = len(np.unique(y))
print(f"{n_labels} labels to predict (y): {np.unique(y)}")
print(f"fMRI data shape (X): {fmri_img.shape}")
print(f"Runs (groups): {np.unique(run)}")
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
8 labels to predict (y): ['bottle' 'cat' 'chair' 'face' 'house' 'scissors' 'scrambledpix' 'shoe']
fMRI data shape (X): (40, 64, 64, 864)
Runs (groups): [ 0 1 2 3 4 5 6 7 8 9 10 11]
Preprocessing¶
As we can see, the fMRI data is a 4D image with shape (40, 64, 64, 864). Here 40x64x64 are the dimensions of the 3D brain image and 864 is the number of brain volumes acquired while visual stimuli were presented, each corresponding to one of the 8 labels we selected above.
Decoder can convert this 4D image to a 2D numpy
array where each row corresponds to a trial and each column corresponds to a
voxel. In addition, it can also do several other things like masking,
smoothing, standardizing the data etc. depending on your requirements.
Under the hood, Decoder uses
NiftiMasker to do all these operations. So here we
will demonstrate this by directly using the
NiftiMasker. Specifically, we will use it to:
1. only keep the data from the Ventral Temporal cortex by providing the
mask image (in Decoder this is done by
providing the mask image in the mask parameter).
2. standardize the data by z-scoring it such that the data is scaled to
have zero mean and unit variance across trials (in
Decoder
this is done by setting the standardize
parameter to "zscore_sample").
from nilearn.maskers import NiftiMasker
masker = NiftiMasker(mask_img=mask_vt, standardize="zscore_sample", verbose=1)
Convert the multi-class labels to binary labels¶
The Decoder converts multi-class classification
problem to N one-vs-others binary classification problems by default (where N
is the number of unique labels)
The advantage of this approach is its interpretability. Once we are done with training and cross-validating, we will have N area-under receiver operating characteristic curve (AU-ROC) scores, one for each label. This will give us an insight into which labels (and the corresponding cognitive domains) are easier to predict and are hence well differentiated relative to the others in the brain.
In addition, we will also have access to the classifier coefficients for each label. These can be further used to understand the importance of each voxel for each corresponding cognitive domain.
In this example we have N = 8 unique labels and we will use Scikit-Learn’s
LabelBinarizer to do this conversion.
from matplotlib import pyplot as plt
from sklearn.preprocessing import LabelBinarizer
label_binarizer = LabelBinarizer(pos_label=1, neg_label=-1)
y_binary = label_binarizer.fit_transform(y)
Let’s plot the labels to understand the conversion
from matplotlib.colors import ListedColormap
from sklearn.preprocessing import LabelEncoder
# create a copy of y_binary and manipulate it just for plotting
y_binary_ = y_binary.copy()
for col in range(y_binary_.shape[1]):
y_binary_[np.where(y_binary_[:, col] == 1), col] = col
fig, (ax_binary, ax_multi) = plt.subplots(
2, gridspec_kw={"height_ratios": [10, 1.5]}, figsize=(12, 2)
)
cmap = ListedColormap(["white", *list(plt.cm.tab10.colors)[:n_labels]])
binary_plt = ax_binary.imshow(
y_binary_.T,
aspect="auto",
cmap=cmap,
interpolation="nearest",
origin="lower",
)
ax_binary.set_xticks([])
ax_binary.set_yticks([])
ax_binary.set_ylabel("One-vs-Others")
# encode the original labels for plotting
label_multi = LabelEncoder()
y_multi = label_multi.fit_transform(y)
y_multi = y_multi.reshape(1, -1)
cmap = ListedColormap(list(plt.cm.tab10.colors)[:n_labels])
multi_plt = ax_multi.imshow(
y_multi,
aspect="auto",
interpolation="nearest",
cmap=cmap,
)
ax_multi.set_yticks([])
ax_multi.set_xlabel("Original trial sequence")
cbar = fig.colorbar(multi_plt, ax=[ax_binary, ax_multi])
cbar.set_ticks(np.arange(1 + len(label_multi.classes_)))
cbar.set_ticklabels([*label_multi.classes_, "all others"])
plt.show()
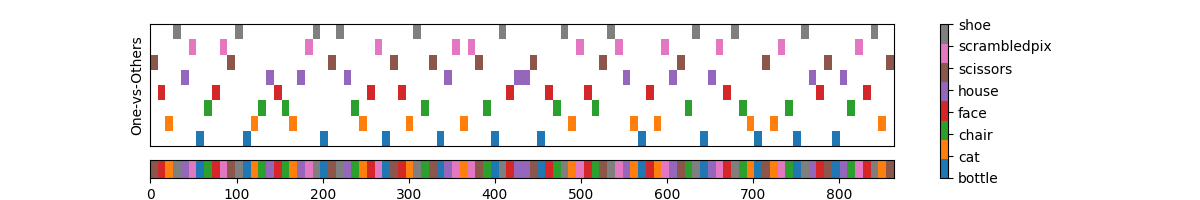
So at the bottom we have the original presentation sequence of the selected trials and at the top we have the labels in the one-vs-others format.
Each row corresponds to a one-vs-others binary classification problem. For example, the first row from the bottom corresponds to the binary classification problem of predicting the label “bottle” vs. all other labels and so on. Later we will train a classifier for each row and calculate the AU-ROC score for each row.
Feature selection¶
After preprocessing the provided fMRI data, the
Decoder performs a univariate feature selection on
the voxels of the brain volume. It uses Scikit-Learn’s
SelectPercentile with
f_classif to calculate ANOVA F-scores for
each voxel and to only keep the ones that have highest 20 percentile scores,
by default. This selection threshold can be changed using the
screening_percentile parameter.
These 20 percentile voxels are with respect to the volume of the standard MNI152 brain template. Furthermore, if the provided mask image has less voxels than the selected percentile, all voxels in the mask are used.
Also note that these top 20 percentile voxels are selected based on training set and then these selected voxels are picked for the test set too for each train-test split.
For simplicity we will just keep all (100 percentile) voxels in this example.
from sklearn.feature_selection import SelectPercentile, f_classif
screening_percentile = 100
feature_selector = SelectPercentile(f_classif, percentile=screening_percentile)
Hyperparameter optimization¶
The Decoder also performs hyperparameter tuning.
How this is done depends on the estimator used.
For the support vector classifiers (known as SVC, and used by setting
estimator="svc" or "svc_l1" or "svc_l2"), the score from the
best performing regularization hyperparameter (C) for each train-test
split is picked.
For all classifiers other than SVC, the hyperparameter tuning is done using
the <estimator_name>CV classes from Scikit-Learn. This essentially means
that the hyperparameters are optimized using an internal cross-validation on
the training data.
In addition, the parameter grids that are used for hyperparameter tuning
by Decoder are also different from the default
Scikit-Learn parameter grids for the corresponding <estimator_name>CV
objects.
For simplicity, let’s use Scikit-Learn’s
LogisticRegressionCV with custom parameter
grid (via Cs parameter) as used in Nilearn’s
Decoder.
from sklearn.linear_model import LogisticRegressionCV
classifier = LogisticRegressionCV(
penalty="l2",
solver="liblinear",
Cs=np.geomspace(1e-3, 1e4, 8),
refit=True,
verbose=1,
)
Train and cross-validate via an Scikit-Learn pipeline¶
Now let’s put all the pieces together to train and cross-validate. The
Decoder uses a leave-one-group-out
cross-validation scheme by default in cases where groups are defined. In our
example a group is a run, so we will use Scikit-Learn’s
LeaveOneGroupOut
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import LeaveOneGroupOut
logo_cv = LeaveOneGroupOut()
# Transform fMRI data into a 2D numpy array and standardize it with the masker
X = masker.fit_transform(fmri_img)
print(f"fMRI data shape after masking: {X.shape}")
# So now we have a 2D numpy array of shape (864, 464) where each row
# corresponds to a trial and each column corresponds to a feature
# (voxel in the Ventral Temporal cortex).
# Loop over each CV split and each class vs. rest binary classification
# problems (number of classification problems = n_labels)
scores_sklearn = []
for klass in range(n_labels):
for train, test in logo_cv.split(X, y, groups=run):
# separate train and test events in the data
X_train, X_test = X[train], X[test]
# separate labels for train and test events for a given class vs. rest
# problem
y_train, y_test = y_binary[train, klass], y_binary[test, klass]
# select the voxels by fitting feature selector on training data
X_train = feature_selector.fit_transform(X_train, y_train)
# pick the same voxels in the test data
X_test = feature_selector.transform(X_test)
# fit the classifier on the training data
classifier.fit(X_train, y_train)
# predict the labels on the test data
pred = classifier.predict_proba(X_test)
# calculate the ROC AUC score
score = roc_auc_score(y_test, pred[:, 1])
scores_sklearn.append(score)
[NiftiMasker.wrapped] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ee890b340>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_understand_decoder.py:254: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[NiftiMasker.wrapped] Resampling mask
[NiftiMasker.wrapped] Finished fit
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ee890b340>
[NiftiMasker.wrapped] Extracting region signals
[NiftiMasker.wrapped] Cleaning extracted signals
fMRI data shape after masking: (864, 464)
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.0s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.8s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 1.9s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.2s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
[LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][LibLinear][Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.1s finished
Decode via the Decoder¶
All these steps can be done in a few lines and made faster via parallel
processing using the n_jobs parameter in
Decoder.
from nilearn.decoding import Decoder
decoder = Decoder(
estimator="logistic_l2",
mask=mask_vt,
standardize="zscore_sample",
n_jobs=n_labels,
cv=logo_cv,
screening_percentile=screening_percentile,
scoring="roc_auc_ovr",
verbose=1,
)
decoder.fit(fmri_img, y, groups=run)
scores_nilearn = np.concatenate(list(decoder.cv_scores_.values()))
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/subj2/mask4_vt.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ee890b340>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_understand_decoder.py:305: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ee890b340>
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Parallel(n_jobs=8)]: Using backend LokyBackend with 8 concurrent workers.
[Parallel(n_jobs=8)]: Done 25 tasks | elapsed: 35.8s
[Parallel(n_jobs=8)]: Done 96 out of 96 | elapsed: 1.6min finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Compare the results¶
Let’s compare the results from the Scikit-Learn pipeline and the Nilearn decoder.
print("Nilearn mean AU-ROC score", np.mean(scores_nilearn))
print("Scikit-Learn mean AU-ROC score", np.mean(scores_sklearn))
Nilearn mean AU-ROC score 0.9073798500881832
Scikit-Learn mean AU-ROC score 0.9073798500881832
As we can see, the mean AU-ROC scores from the Scikit-Learn pipeline and
Nilearn’s Decoder are identical.
The advantage of using Nilearn’s Decoder is
that it does all these steps under the hood and provides a simple interface
to train, cross-validate and predict on new data, while also parallelizing
the computations to make the cross-validation faster. It also organizes the
results in a structured way that can be easily accessed and analyzed.
Total running time of the script: (5 minutes 20.018 seconds)
Estimated memory usage: 1218 MB