Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Statistical testing of a second-level analysis¶
Perform a one-sample t-test on a bunch of images (a.k.a. second-level analysis in fMRI) and threshold the resulting statistical map.
This example is based on the so-called localizer dataset. It shows activation related to a mental computation task, as opposed to narrative sentence reading/listening.
Prepare some images for a simple t test¶
This is a simple manually performed second level analysis.
from nilearn import datasets
n_samples = 20
localizer_dataset = datasets.fetch_localizer_calculation_task(
n_subjects=n_samples,
)
[fetch_localizer_calculation_task] Dataset found in /home/remi-gau/nilearn_data/brainomics_localizer
Get the set of individual statstical maps (contrast estimates)
Perform the second level analysis¶
First, we define a design matrix for the model. As the model is trivial (one-sample test), the design matrix is just one column with ones.
import pandas as pd
design_matrix = pd.DataFrame([1] * n_samples, columns=["intercept"])
Next, we specify and estimate the model.
from nilearn.glm.second_level import SecondLevelModel
second_level_model = SecondLevelModel(n_jobs=2, verbose=1).fit(
cmap_filenames, design_matrix=design_matrix
)
[SecondLevelModel.fit] Fitting second level model. Take a deep breath.
[SecondLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at 0x7587a4f1cb20>
[SecondLevelModel.fit] Computing mask
[SecondLevelModel.fit] Resampling mask
[SecondLevelModel.fit] Finished fit
[SecondLevelModel.fit]
Computation of second level model done in 0.31 seconds.
Compute the only possible contrast: the one-sample test. Since there is only one possible contrast, we don’t need to specify it in detail.
z_map = second_level_model.compute_contrast(output_type="z_score")
[SecondLevelModel.compute_contrast] Loading data from <nibabel.nifti1.Nifti1Image object at 0x7587eab1c430>
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
Threshold the resulting map without multiple comparisons correction, abs(z) > 3.29 (equivalent to p < 0.001), cluster size > 10 voxels.
from nilearn.image import threshold_img
threshold_img(
z_map,
threshold=3.29,
cluster_threshold=10,
two_sided=True,
)
<nibabel.nifti1.Nifti1Image object at 0x7587a4f1cca0>
This is equivalent to thresholding a z-statistic image with a false positive rate < .001, cluster size > 10 voxels.
from nilearn.glm import threshold_stats_img
thresholded_map1, threshold1 = threshold_stats_img(
z_map,
alpha=0.001,
height_control="fpr",
cluster_threshold=10,
two_sided=True,
)
Now use FDR <.05 (False Discovery Rate) and no cluster-level threshold.
thresholded_map2, threshold2 = threshold_stats_img(
z_map, alpha=0.05, height_control="fdr"
)
print(f"The FDR=.05 threshold is {threshold2:.3g}")
The FDR=.05 threshold is 2.37
Now use FWER <.05 (Family-Wise Error Rate) and no cluster-level threshold. As the data has not been intensively smoothed, we can use a simple Bonferroni correction.
thresholded_map3, threshold3 = threshold_stats_img(
z_map, alpha=0.05, height_control="bonferroni"
)
print(f"The p<.05 Bonferroni-corrected threshold is {threshold3:.3g}")
The p<.05 Bonferroni-corrected threshold is 4.88
Visualize the results¶
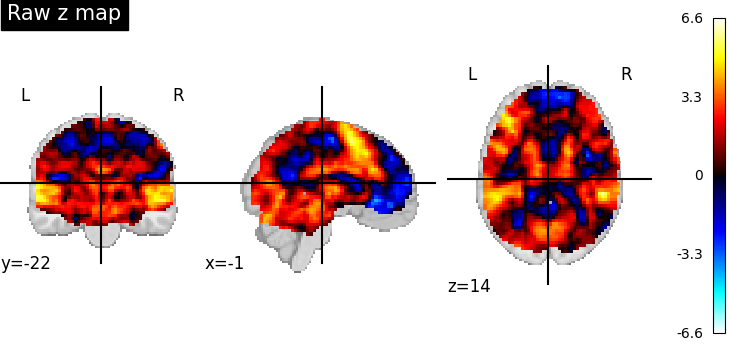
First, the unthresholded map.
from nilearn import plotting
display = plotting.plot_stat_map(z_map, title="Raw z map")
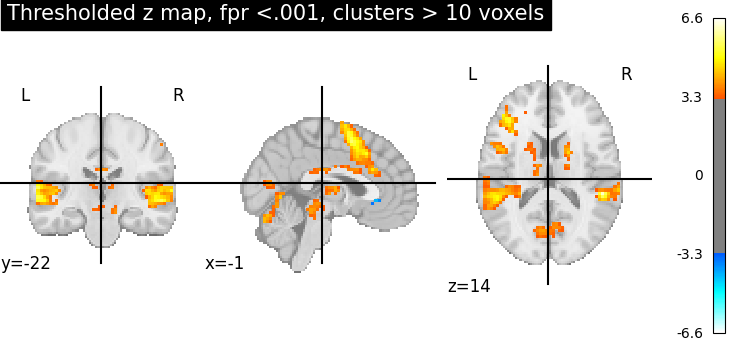
Second, the p<.001 uncorrected-thresholded map (with only clusters > 10 voxels).
plotting.plot_stat_map(
thresholded_map1,
cut_coords=display.cut_coords,
threshold=threshold1,
title="Thresholded z map, fpr <.001, clusters > 10 voxels",
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7587eb3e8070>
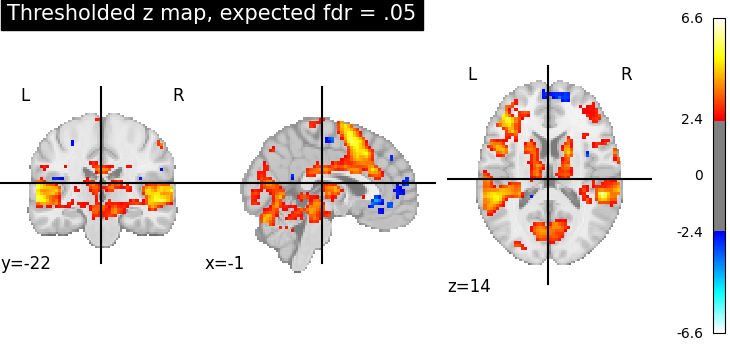
Third, the fdr-thresholded map.
plotting.plot_stat_map(
thresholded_map2,
cut_coords=display.cut_coords,
title="Thresholded z map, expected fdr = .05",
threshold=threshold2,
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7587ebd4bb20>
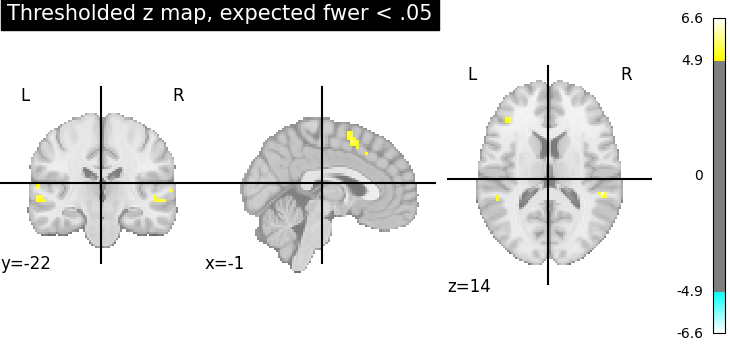
Fourth, the Bonferroni-thresholded map.
plotting.plot_stat_map(
thresholded_map3,
cut_coords=display.cut_coords,
title="Thresholded z map, expected fwer < .05",
threshold=threshold3,
)
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x75876ac76f80>
These different thresholds correspond to different statistical guarantees: in the FWER-corrected image there is only a probability smaller than .05 of observing any false positive voxel. In the FDR-corrected image, 5% of the voxels found are likely to be false positive. In the uncorrected image, one expects a few tens of false positive voxels.
Total running time of the script: (0 minutes 16.254 seconds)
Estimated memory usage: 98 MB