Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.connectome.ConnectivityMeasure¶
- class nilearn.connectome.ConnectivityMeasure(cov_estimator=None, kind='covariance', vectorize=False, discard_diagonal=False, standardize=True, verbose=0)[source]¶
A class that computes different kinds of functional connectivity matrices.
Added in Nilearn 0.2.
- Parameters:
- cov_estimatorestimator object, default=LedoitWolf(store_precision=False)
The covariance estimator. This implies that correlations are slightly shrunk towards zero compared to a maximum-likelihood estimate
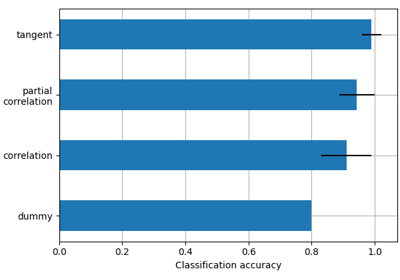
- kind{“covariance”, “correlation”, “partial correlation”, “tangent”, “precision”}, default=’covariance’
The matrix kind. For the use of “tangent” see Varoquaux et al.[1].
- vectorize
bool, default=False If True, connectivity matrices are reshaped into 1D arrays and only their flattened lower triangular parts are returned.
- discard_diagonal
bool, default=False If True, vectorized connectivity coefficients do not include the matrices diagonal elements. Used only when vectorize is set to True.
- standardizeany of: ‘zscore_sample’, ‘zscore’, ‘psc’, True, False or None; default=True
Strategy to standardize the signal:
'zscore_sample': The signal is z-scored. Timeseries are shifted to zero mean and scaled to unit variance. Uses sample std.'zscore': The signal is z-scored. Timeseries are shifted to zero mean and scaled to unit variance. Uses population std by calling defaultnumpy.stdwith N -ddof=0.Deprecated since Nilearn 0.10.1: This option will be removed in Nilearn version 0.14.0. Use
zscore_sampleinstead.'psc': Timeseries are shifted to zero mean value and scaled to percent signal change (as compared to original mean signal).True: The signal is z-scored (same as option zscore). Timeseries are shifted to zero mean and scaled to unit variance.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Truewill be replaced by'zscore_sample'.False: Do not standardize the data.Deprecated since Nilearn 0.13.0: In nilearn version 0.15.0,
Falsewill be replaced byNone.
Deprecated since Nilearn 0.10.1: The default will be changed to
'zscore_sample'and'zscore'will be removed in in version 0.14.0.Deprecated since Nilearn 0.15.0dev: The default will be changed to
'zscore_sample'in version 0.15.0.Note
Added to control passing value to standardize of
signal.cleanto call new behavior since passing “zscore” or True (default) is deprecated. This parameter will be changed to “zscore_sample” in version 0.14 and removed in version 0.15.- verbose
boolorint, default=0 Verbosity level (
0orFalsemeans no message).
- Attributes:
- cov_estimator_estimator object, default=None
A new covariance estimator with the same parameters as cov_estimator. If
Noneis passed, defaults toLedoitWolf(store_precision=False).- mean_numpy.ndarray
The mean connectivity matrix across subjects. For ‘tangent’ kind, it is the geometric mean of covariances (a group covariance matrix that captures information from both correlation and partial correlation matrices). For other values for “kind”, it is the mean of the corresponding matrices
- n_features_in_
int Number of features seen during fit.
- whitening_numpy.ndarray or None
The inverted square-rooted geometric mean of the covariance matrices. Only set when for
kind=="tangent"
References
- __init__(cov_estimator=None, kind='covariance', vectorize=False, discard_diagonal=False, standardize=True, verbose=0)[source]¶
- fit(X, y=None)[source]¶
Fit the covariance estimator to the given time series for each subject.
- Parameters:
- Xiterable of
numpy.ndarrayeach of shape (n_samples, n_features) Each
numpy.ndarrayrepresents a subject’s time series. The number of samples may differ from one subject to another.- yNone
This parameter is unused. It is solely included for scikit-learn compatibility.
- Xiterable of
- Returns:
- selfConnectivityMatrix instance
The object itself. Useful for chaining operations.
- fit_transform(X, y=None, confounds=None)[source]¶
Fit the covariance estimator to the given time series for each subject. Then apply transform to covariance matrices for the chosen kind.
- Parameters:
- Xiterable of
numpy.ndarrayeach of shape (n_samples, n_features) Each
numpy.ndarrayrepresents a subject’s time series. The number of samples may differ from one subject to another.- yNone
This parameter is unused. It is solely included for scikit-learn compatibility.
- confoundsnp.ndarray with shape (n_samples) or (n_samples, n_confounds), or pandas DataFrame, default=None
Confounds to be cleaned on the vectorized matrices. Only takes into effect when vetorize=True. This parameter is passed to signal.clean. Please see the related documentation for details.
- Xiterable of
- Returns:
- outputnumpy.ndarray, shape (n_subjects, n_features, n_features) or (n_subjects, n_features * (n_features + 1) / 2) if vectorize is set to True.
The transformed individual connectivities, as matrices or vectors. Vectors are cleaned when vectorize=True and confounds are provided.
- get_metadata_routing()¶
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- inverse_transform(connectivities, diagonal=None)[source]¶
Return connectivity matrices from connectivities, vectorized or not.
If kind is ‘tangent’, the covariance matrices are reconstructed.
- Parameters:
- connectivities
listof n_subjects numpy.ndarray with shapes (n_features, n_features) or (n_features * (n_features + 1) / 2,) or ((n_features - 1) * n_features / 2,) Connectivities of each subject, vectorized or not.
- diagonalnumpy.ndarray, shape (n_subjects, n_features), default=None
The diagonals of the connectivity matrices.
- connectivities
- Returns:
- outputnumpy.ndarray, shape (n_subjects, n_features, n_features)
The corresponding connectivity matrices. If kind is ‘correlation’/ ‘partial correlation’, the correlation/partial correlation matrices are returned. If kind is ‘tangent’, the covariance matrices are reconstructed.
- set_inverse_transform_request(*, connectivities='$UNCHANGED$', diagonal='$UNCHANGED$')¶
Configure whether metadata should be requested to be passed to the
inverse_transformmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed toinverse_transformif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it toinverse_transform.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- connectivitiesstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
connectivitiesparameter ininverse_transform.- diagonalstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
diagonalparameter ininverse_transform.
- Returns:
- selfobject
The updated object.
- set_output(*, transform=None)[source]¶
Set the output container when
"transform"is called.Warning
This has not been implemented yet.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
- set_transform_request(*, confounds='$UNCHANGED$')¶
Configure whether metadata should be requested to be passed to the
transformmethod.Note that this method is only relevant when this estimator is used as a sub-estimator within a meta-estimator and metadata routing is enabled with
enable_metadata_routing=True(seesklearn.set_config). Please check the User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed totransformif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it totransform.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
- Parameters:
- confoundsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
confoundsparameter intransform.
- Returns:
- selfobject
The updated object.
- transform(X, confounds=None)[source]¶
Apply transform to covariances matrices to get the connectivity matrices for the chosen kind.
- Parameters:
- Xiterable of
numpy.ndarrayeach of shape (n_samples, n_features) Each
numpy.ndarrayrepresents a subject’s time series. The number of samples may differ from one subject to another.- confoundsnumpy.ndarray with shape (n_samples) or (n_samples, n_confounds), default=None
Confounds to be cleaned on the vectorized matrices. Only takes into effect when vetorize=True. This parameter is passed to signal.clean. Please see the related documentation for details.
- Xiterable of
- Returns:
- outputnumpy.ndarray, shape (n_subjects, n_features, n_features) or (n_subjects, n_features * (n_features + 1) / 2) if vectorize is set to True.
The transformed individual connectivities, as matrices or vectors. Vectors are cleaned when vectorize=True and confounds are provided.
Examples using nilearn.connectome.ConnectivityMeasure¶
Classification of age groups using functional connectivity
Comparing connectomes on different reference atlases
Extracting signals of a probabilistic atlas of functional regions
Regions extraction using dictionary learning and functional connectomes