Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Basic Atlas plotting¶
Plot the regions of reference atlases.
Check the list of atlases to know which ones are shipped with Nilearn.
Retrieving the atlas data¶
from nilearn import datasets
dataset_ho = datasets.fetch_atlas_harvard_oxford("cort-maxprob-thr25-2mm")
atlas_ho_filename = dataset_ho.filename
print(f"Atlas ROIs are located at: {atlas_ho_filename}")
dataset_ju = datasets.fetch_atlas_juelich("maxprob-thr0-1mm")
atlas_ju_filename = dataset_ju.filename
print(f"Atlas ROIs are located at: {atlas_ju_filename}")
[fetch_atlas_harvard_oxford] Dataset created in /home/runner/nilearn_data/fsl
[fetch_atlas_harvard_oxford] Downloading data from
https://www.nitrc.org/frs/download.php/9902/HarvardOxford.tgz ...
[fetch_atlas_harvard_oxford] ...done. (1 seconds, 0 min)
[fetch_atlas_harvard_oxford] Extracting data from
/home/runner/nilearn_data/fsl/5c734f16e50cc772ef593cab9bb3137b/HarvardOxford.tgz
...
[fetch_atlas_harvard_oxford] .. done.
Atlas ROIs are located at: /home/runner/nilearn_data/fsl/data/atlases/HarvardOxford/HarvardOxford-cort-maxprob-thr25-2mm.nii.gz
[fetch_atlas_juelich] Dataset found in /home/runner/nilearn_data/fsl
[fetch_atlas_juelich] Downloading data from
https://www.nitrc.org/frs/download.php/12096/Juelich.tgz ...
[fetch_atlas_juelich] ...done. (1 seconds, 0 min)
[fetch_atlas_juelich] Extracting data from
/home/runner/nilearn_data/fsl/7e62e7e7fcc4d6e1428b6b2cb48f7a7c/Juelich.tgz...
[fetch_atlas_juelich] .. done.
Atlas ROIs are located at: /home/runner/nilearn_data/fsl/data/atlases/Juelich/Juelich-maxprob-thr0-1mm.nii.gz
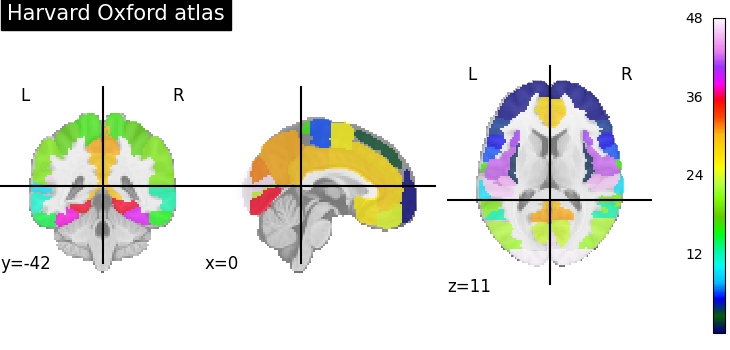
Visualizing the Harvard-Oxford atlas¶
from nilearn.plotting import plot_roi, show
plot_roi(atlas_ho_filename, title="Harvard Oxford atlas")
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0cd651b0>
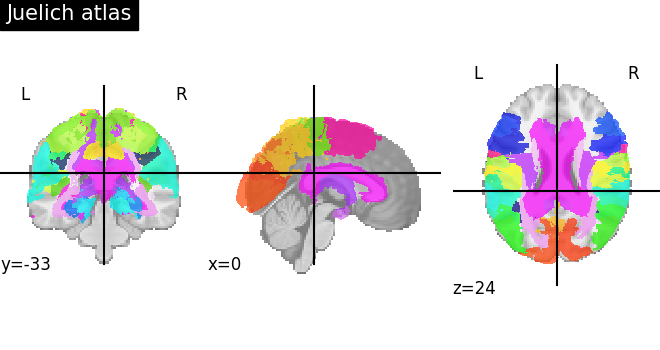
Visualizing the Juelich atlas¶
plot_roi(atlas_ju_filename, title="Juelich atlas")
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee8c852a0>
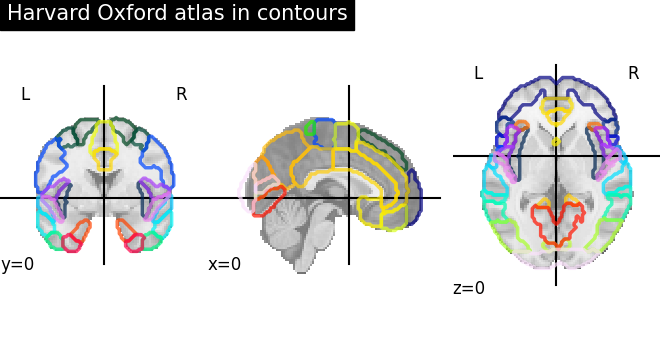
Visualizing the Harvard-Oxford atlas with contours¶
plot_roi(
atlas_ho_filename,
view_type="contours",
title="Harvard Oxford atlas in contours",
)
show()
/home/runner/work/nilearn/nilearn/examples/01_plotting/plot_atlas.py:47: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
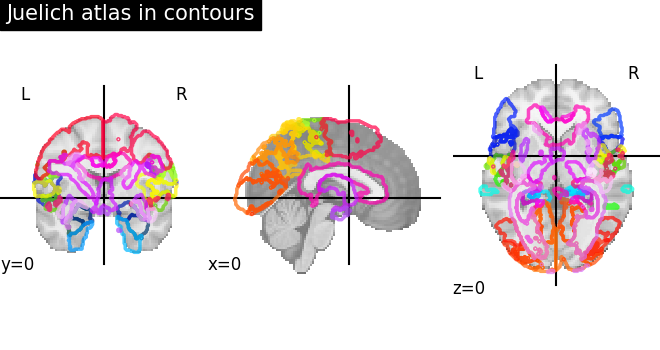
Visualizing the Juelich atlas with contours¶
plot_roi(
atlas_ju_filename, view_type="contours", title="Juelich atlas in contours"
)
show()
/home/runner/work/nilearn/nilearn/examples/01_plotting/plot_atlas.py:55: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Visualizing an atlas with its own colormap¶
Some atlases come with a look-up table that determines the color to use to represent each of its regions.
You can pass this look-up table
as a pandas dataframe to the cmap argument
to use its colormap.
Look-up table format
The look-up table must be formatted according to the BIDS standard.
and that the colors must be in color column using hexadecimal values.
If an invalid look-up table is passed,
a warning will be thrown and the plot_roi function
will fall back to using its default colormap.
Here we are using the Yeo atlas that comes with a predefined colormap.
dataset_yeo = datasets.fetch_atlas_yeo_2011(n_networks=17)
print(dataset_yeo.lut)
[fetch_atlas_yeo_2011] Dataset created in /home/runner/nilearn_data/yeo_2011
[fetch_atlas_yeo_2011] Downloading data from
ftp://surfer.nmr.mgh.harvard.edu/pub/data/Yeo_JNeurophysiol11_MNI152.zip ...
[fetch_atlas_yeo_2011] ...done. (2 seconds, 0 min)
[fetch_atlas_yeo_2011] Extracting data from
/home/runner/nilearn_data/yeo_2011/622858b56913b19ae0865d9fa8ad47cc/Yeo_JNeuroph
ysiol11_MNI152.zip...
[fetch_atlas_yeo_2011] .. done.
index name color
0 0 Background #000000
1 1 17Networks_1 #781286
2 2 17Networks_2 #ff0000
3 3 17Networks_3 #4682b4
4 4 17Networks_4 #2acca4
5 5 17Networks_5 #4a9b3c
6 6 17Networks_6 #00760e
7 7 17Networks_7 #c43afa
8 8 17Networks_8 #ff98d5
9 9 17Networks_9 #dcf8a4
10 10 17Networks_10 #7a8732
11 11 17Networks_11 #778cb0
12 12 17Networks_12 #e69422
13 13 17Networks_13 #87324a
14 14 17Networks_14 #0c30ff
15 15 17Networks_15 #000082
16 16 17Networks_16 #ffff00
17 17 17Networks_17 #cd3e4e
Let’s compare the atlas with the default colormap and its own colormap.
plot_roi(
dataset_yeo.maps,
title="Yeo atlas",
colorbar=True,
)
plot_roi(
dataset_yeo.maps,
title="Yeo atlas with its own colors",
cmap=dataset_yeo.lut,
colorbar=True,
)
show()
/home/runner/work/nilearn/nilearn/examples/01_plotting/plot_atlas.py:101: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (0 minutes 58.434 seconds)
Estimated memory usage: 749 MB

