Note
This page is a reference documentation. It only explains the function signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.image.math_img¶
- nilearn.image.math_img(formula, copy_header_from=None, **imgs)[source]¶
Interpret a numpy based string formula using niimg in named parameters.
Added in Nilearn 0.2.3.
- Parameters:
- formula
str The mathematical formula to apply to image internal data. It can use numpy imported as ‘np’.
- copy_header_from
stror None, default=None Takes the variable name of one of the images in the formula. The header of this image will be copied to the result of the formula. Note that the result image and the image to copy the header from, should have the same number of dimensions. If None, the default
Nifti1Headeris used.Ignored for
SurfaceImage.Added in Nilearn 0.10.4.
- imgsimages (
Nifti1Imageor file names orSurfaceImageobject) Keyword arguments corresponding to the variables in the formula as images. All input images should have the same ‘geometry’:
shape and affine for volume data
mesh (coordinates and faces) for surface data
- formula
- Returns:
Nifti1ImageorSurfaceImageobjectResult of the formula as an image. Note that the dimension of the result image can be smaller than the input image. For volume image input, the affine is the same as the input images. For surface image input, the mesh is the same as the input images.
See also
nilearn.image.mean_imgTo simply compute the mean of multiple images
Notes
This function is the Python equivalent of ImCal in SPM or fslmaths in FSL.
Examples
Let’s load an image using nilearn datasets module:
>>> from nilearn import datasets >>> anatomical_image = datasets.load_mni152_template()
Now we can use any numpy function on this image:
>>> from nilearn.image import math_img >>> log_img = math_img("np.log(img)", img=anatomical_image)
We can also apply mathematical operations on several images:
>>> result_img = math_img("img1 + img2", ... img1=anatomical_image, img2=log_img)
The result image will have the same shape and affine as the input images; but might have different header information, specifically the TR value, see #2645.
Added in Nilearn 0.10.4.
We can also copy the header from one of the input images using
copy_header_from:>>> result_img_with_header = math_img("img1 + img2", ... img1=anatomical_image, img2=log_img, ... copy_header_from="img1")
Examples using nilearn.image.math_img¶
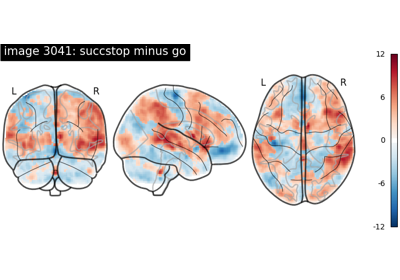
NeuroVault meta-analysis of stop-go paradigm studies