Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Voxel-Based Morphometry on OASIS dataset¶
This example uses voxel-based morphometry (VBM) to study the relationship between aging, sex, and gray matter density.
The data come from the OASIS project. If you use it, you need to agree with the data usage agreement available on the website.
It has been run through a standard VBM pipeline (using SPM8 and NewSegment) to create VBM maps, which we study here.
VBM analysis of aging¶
We run a standard GLM analysis to study the association between age and gray matter density from the VBM data. We use only 100 subjects from the OASIS dataset to limit the memory usage.
Note that more power would be obtained from using a larger sample of subjects.
See also
For more information see the dataset description.
Load Oasis dataset¶
from nilearn.datasets import (
fetch_icbm152_2009,
fetch_icbm152_brain_gm_mask,
fetch_oasis_vbm,
)
from nilearn.plotting import plot_design_matrix, plot_stat_map
n_subjects = 100 # more subjects requires more memory
oasis_dataset = fetch_oasis_vbm(
n_subjects=n_subjects,
)
gray_matter_map_filenames = oasis_dataset.gray_matter_maps
age = oasis_dataset.ext_vars["age"].astype(float)
[fetch_oasis_vbm] Dataset found in /home/runner/nilearn_data/oasis1
Sex is encoded as ‘M’ or ‘F’. Hence, we make it a binary variable.
sex = oasis_dataset.ext_vars["mf"] == "F"
Print basic information on the dataset.
print(
"First gray-matter anatomy image (3D) is located at: "
f"{oasis_dataset.gray_matter_maps[0]}"
)
print(
"First white-matter anatomy image (3D) is located at: "
f"{oasis_dataset.white_matter_maps[0]}"
)
First gray-matter anatomy image (3D) is located at: /home/runner/nilearn_data/oasis1/OAS1_0001_MR1/mwrc1OAS1_0001_MR1_mpr_anon_fslswapdim_bet.nii.gz
First white-matter anatomy image (3D) is located at: /home/runner/nilearn_data/oasis1/OAS1_0001_MR1/mwrc2OAS1_0001_MR1_mpr_anon_fslswapdim_bet.nii.gz
Get a mask image: A mask of the cortex of the ICBM template.
[fetch_icbm152_brain_gm_mask] Dataset found in
/home/runner/nilearn_data/icbm152_2009
Resample the mask, since this mask has a different resolution.
from nilearn.image import resample_to_img
mask_img = resample_to_img(
gm_mask,
gray_matter_map_filenames[0],
interpolation="nearest",
)
Analyze data¶
First, we create an adequate design matrix with three columns: ‘age’, ‘sex’, and ‘intercept’.
import numpy as np
import pandas as pd
intercept = np.ones(n_subjects)
design_matrix = pd.DataFrame(
np.vstack((age, sex, intercept)).T,
columns=["age", "sex", "intercept"],
)
from matplotlib import pyplot as plt
Let’s plot the design matrix.
fig, ax1 = plt.subplots(1, 1, figsize=(4, 8))
ax = plot_design_matrix(design_matrix, axes=ax1)
ax.set_ylabel("maps")
fig.suptitle("Second level design matrix")
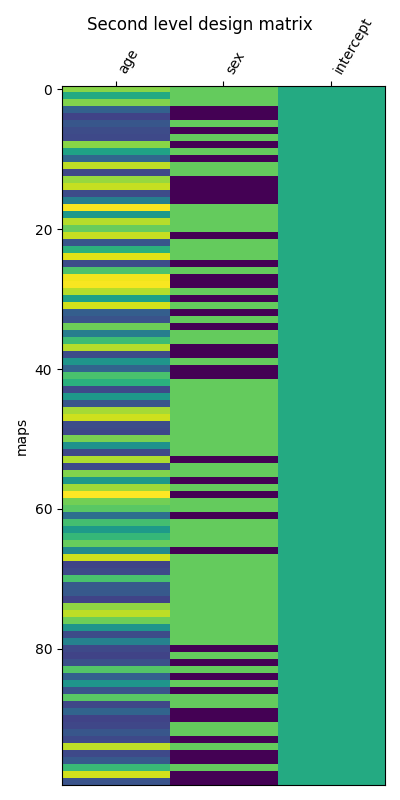
Text(0.5, 0.98, 'Second level design matrix')
Next, we specify and fit the second-level model when loading the data and also smooth a little bit to improve statistical behavior.
from nilearn.glm.second_level import SecondLevelModel
second_level_model = SecondLevelModel(
smoothing_fwhm=2.0,
mask_img=mask_img,
n_jobs=2,
minimize_memory=False,
verbose=1,
)
second_level_model.fit(
gray_matter_map_filenames,
design_matrix=design_matrix,
)
[SecondLevelModel.fit] Fitting second level model. Take a deep breath.
[SecondLevelModel.fit] Loading mask from <nibabel.nifti1.Nifti1Image object at
0x7f1eb1e80e50>
[SecondLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef0253b50>
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_oasis.py:114: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[SecondLevelModel.fit] Resampling mask
[SecondLevelModel.fit] Finished fit
[SecondLevelModel.fit]
Computation of second level model done in 0.51 seconds.
Estimating the contrast is very simple. We can just provide the column name of the design matrix.
z_map = second_level_model.compute_contrast(
second_level_contrast=[1, 0, 0],
output_type="z_score",
)
[SecondLevelModel.compute_contrast] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f1ef0585cc0>
[SecondLevelModel.compute_contrast] Smoothing images
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
View results¶
We threshold the second level contrast at FDR-corrected p < 0.05 and plot it.
from nilearn.glm import threshold_stats_img
from nilearn.plotting import show
_, threshold = threshold_stats_img(z_map, alpha=0.05, height_control="fdr")
print(f"The FDR=.05-corrected threshold is: {threshold:03g}")
fig = plt.figure(figsize=(5, 3))
display = plot_stat_map(
z_map,
threshold=threshold,
display_mode="z",
cut_coords=[-4, 26],
figure=fig,
)
fig.suptitle("age effect on gray matter density (FDR = .05)")
show()
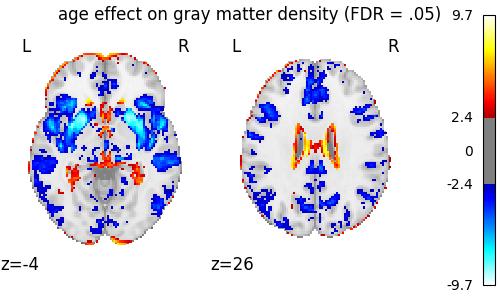
The FDR=.05-corrected threshold is: 2.40175
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_oasis.py:147: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
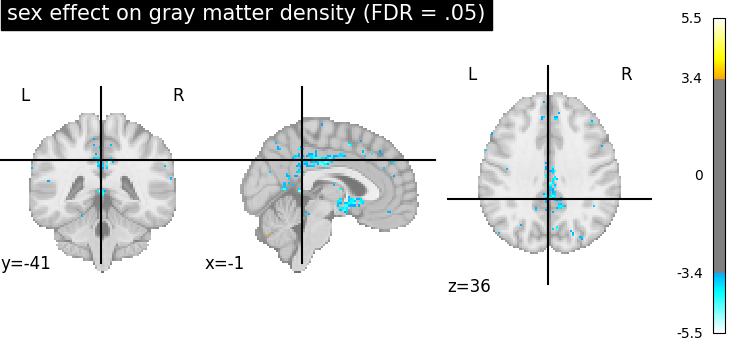
We can also study the effect of sex by computing the contrast, thresholding it and plot the resulting map.
z_map = second_level_model.compute_contrast(
second_level_contrast="sex",
output_type="z_score",
)
_, threshold = threshold_stats_img(z_map, alpha=0.05, height_control="fdr")
plot_stat_map(
z_map,
threshold=threshold,
title="sex effect on gray matter density (FDR = .05)",
)
show()
[SecondLevelModel.compute_contrast] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f1eb1e82530>
[SecondLevelModel.compute_contrast] Smoothing images
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_oasis.py:162: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Note that there does not seem to be any significant effect of sex on gray matter density on that dataset.
Generate a report for the GLM¶
Generate a report and view it.
icbm152_2009 = fetch_icbm152_2009()
report = second_level_model.generate_report(
bg_img=icbm152_2009["t1"],
plot_type="glass",
alpha=0.05,
height_control=None,
)
[fetch_icbm152_2009] Dataset found in /home/runner/nilearn_data/icbm152_2009
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_oasis.py:177: FutureWarning:
From nilearn version>=0.15, the default 'threshold' will be set to 3.090232306167813.
[SecondLevelModel.generate_report] Generating contrast-level figures...
[SecondLevelModel.generate_report] Generating design matrices figures...
[SecondLevelModel.generate_report] Generating contrast matrices figures...
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_oasis.py:177: UserWarning:
No contrast passed during report generation.
Note
The generated report can be:
displayed in a Notebook,
opened in a browser using the
.open_in_browser()method,or saved to a file using the
.save_as_html(output_filepath)method.
Total running time of the script: (0 minutes 40.136 seconds)
Estimated memory usage: 1649 MB