Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Group Sparse inverse covariance for multi-subject connectome¶
This example shows how to estimate a connectome on a group of subjects using the group sparse inverse covariance estimate.
Warning
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
import numpy as np
from nilearn import plotting
n_subjects = 4 # subjects to consider for group-sparse covariance (max: 40)
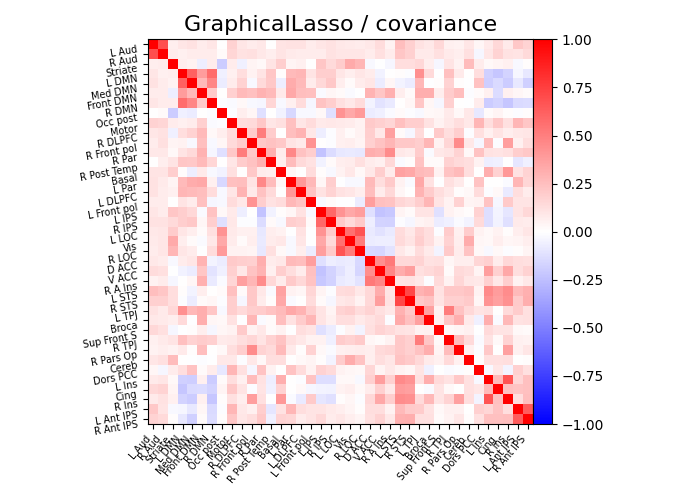
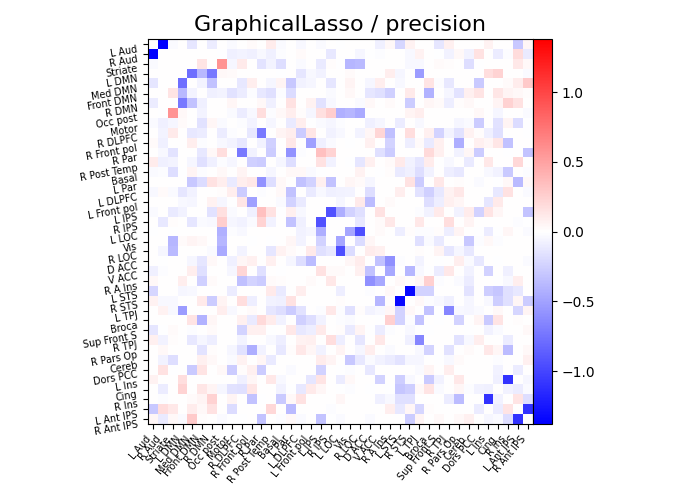
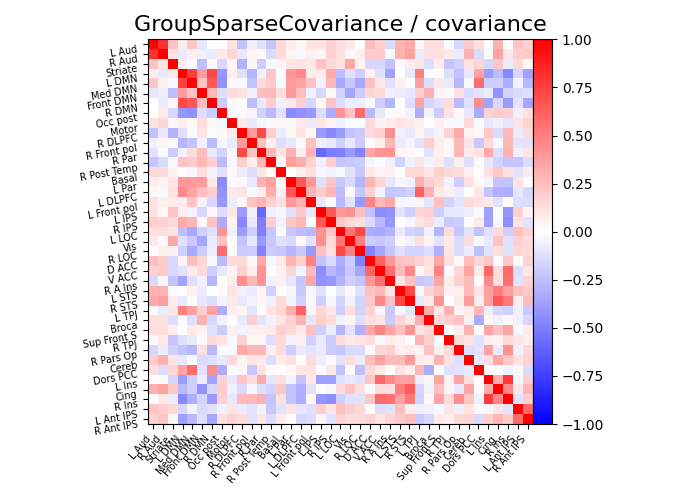
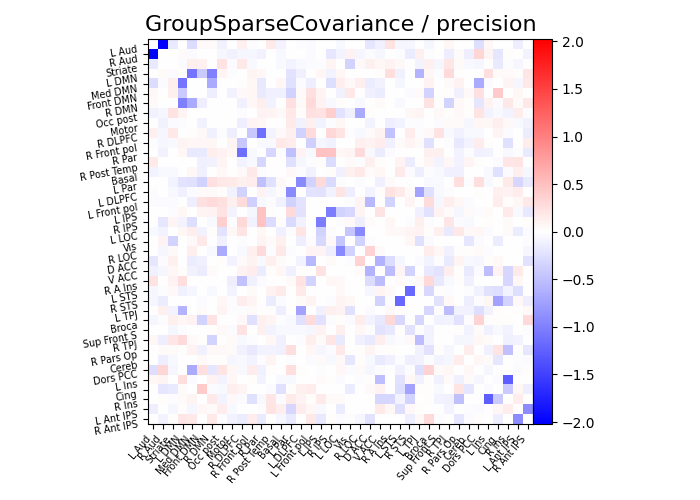
def plot_matrices(cov, prec, title, labels):
"""Plot covariance and precision matrices, for a given processing."""
prec = prec.copy() # avoid side effects
# Put zeros on the diagonal, for graph clarity.
size = prec.shape[0]
prec[list(range(size)), list(range(size))] = 0
span = max(abs(prec.min()), abs(prec.max()))
# Display covariance matrix
plotting.plot_matrix(
cov,
vmin=-1,
vmax=1,
title=f"{title} / covariance",
labels=labels,
)
# Display precision matrix
plotting.plot_matrix(
prec,
vmin=-span,
vmax=span,
title=f"{title} / precision",
labels=labels,
)
Fetching datasets¶
from nilearn.datasets import fetch_atlas_msdl, fetch_development_fmri
msdl_atlas_dataset = fetch_atlas_msdl()
rest_dataset = fetch_development_fmri(n_subjects=n_subjects)
# print basic information on the dataset
print(
f"First subject functional nifti image (4D) is at: {rest_dataset.func[0]}"
)
[fetch_atlas_msdl] Dataset found in /home/remi-gau/nilearn_data/msdl_atlas
[fetch_development_fmri] Dataset found in /home/remi-gau/nilearn_data/development_fmri
[fetch_development_fmri] Dataset found in /home/remi-gau/nilearn_data/development_fmri/development_fmri
[fetch_development_fmri] Dataset found in /home/remi-gau/nilearn_data/development_fmri/development_fmri
First subject functional nifti image (4D) is at: /home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
Extracting region signals¶
from nilearn.maskers import NiftiMapsMasker
masker = NiftiMapsMasker(
msdl_atlas_dataset.maps,
resampling_target="maps",
detrend=True,
high_variance_confounds=True,
low_pass=None,
high_pass=0.01,
t_r=rest_dataset.t_r,
standardize="zscore_sample",
standardize_confounds=True,
memory="nilearn_cache",
memory_level=1,
verbose=1,
)
subject_time_series = []
func_filenames = rest_dataset.func
confound_filenames = rest_dataset.confounds
for func_filename, confound_filename in zip(
func_filenames, confound_filenames, strict=False
):
print(f"Processing file {func_filename}")
region_ts = masker.fit_transform(
func_filename, confounds=confound_filename
)
subject_time_series.append(region_ts)
Processing file /home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
[NiftiMapsMasker.wrapped] Loading regions from '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.image.image.high_variance_confounds...
high_variance_confounds('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
__________________________________________high_variance_confounds - 0.3s, 0.0min
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7587ea174940>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': True,
'keep_masked_maps': False,
'low_pass': None,
'maps_img': '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': 2,
'target_affine': array([[ 4., 0., 0., -78.],
[ 0., 4., 0., -111.],
[ 0., 0., 4., -51.],
[ 0., 0., 0., 1.]]),
'target_shape': (40, 48, 35)}, confounds=[ array([[ 0.174325, ..., -0.048779],
...,
[ 0.044073, ..., 0.155444]], shape=(168, 5)),
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar123_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Resampling images
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/remi-gau/github/nilearn/nilearn/examples/03_connectivity/plot_multi_subject_connectome.py:89: DeprecationWarning: From release 0.14.0, confounds will be standardized using the sample std instead of the population std.
region_ts = masker.fit_transform(
_______________________________________________filter_and_extract - 2.4s, 0.0min
Processing file /home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
[NiftiMapsMasker.wrapped] Loading regions from '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.image.image.high_variance_confounds...
high_variance_confounds('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
__________________________________________high_variance_confounds - 0.3s, 0.0min
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x758791612320>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': True,
'keep_masked_maps': False,
'low_pass': None,
'maps_img': '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': 2,
'target_affine': array([[ 4., 0., 0., -78.],
[ 0., 4., 0., -111.],
[ 0., 0., 4., -51.],
[ 0., 0., 0., 1.]]),
'target_shape': (40, 48, 35)}, confounds=[ array([[-0.151677, ..., -0.057023],
...,
[-0.206928, ..., 0.102714]], shape=(168, 5)),
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar001_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Resampling images
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/remi-gau/github/nilearn/nilearn/examples/03_connectivity/plot_multi_subject_connectome.py:89: DeprecationWarning: From release 0.14.0, confounds will be standardized using the sample std instead of the population std.
region_ts = masker.fit_transform(
_______________________________________________filter_and_extract - 2.4s, 0.0min
Processing file /home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
[NiftiMapsMasker.wrapped] Loading regions from '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.image.image.high_variance_confounds...
high_variance_confounds('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
__________________________________________high_variance_confounds - 0.3s, 0.0min
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7587916127a0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': True,
'keep_masked_maps': False,
'low_pass': None,
'maps_img': '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': 2,
'target_affine': array([[ 4., 0., 0., -78.],
[ 0., 4., 0., -111.],
[ 0., 0., 4., -51.],
[ 0., 0., 0., 1.]]),
'target_shape': (40, 48, 35)}, confounds=[ array([[ 0.127944, ..., -0.087084],
...,
[-0.015679, ..., -0.02587 ]], shape=(168, 5)),
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar002_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Resampling images
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/remi-gau/github/nilearn/nilearn/examples/03_connectivity/plot_multi_subject_connectome.py:89: DeprecationWarning: From release 0.14.0, confounds will be standardized using the sample std instead of the population std.
region_ts = masker.fit_transform(
_______________________________________________filter_and_extract - 2.4s, 0.0min
Processing file /home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz
[NiftiMapsMasker.wrapped] Loading regions from '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii'
[NiftiMapsMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.image.image.high_variance_confounds...
high_variance_confounds('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz')
__________________________________________high_variance_confounds - 0.3s, 0.0min
________________________________________________________________________________
[Memory] Calling nilearn.maskers.base_masker.filter_and_extract...
filter_and_extract('/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
<nilearn.maskers.nifti_maps_masker._ExtractionFunctor object at 0x7587916127a0>, { 'allow_overlap': True,
'clean_args': None,
'clean_kwargs': {},
'cmap': 'CMRmap_r',
'detrend': True,
'dtype': None,
'high_pass': 0.01,
'high_variance_confounds': True,
'keep_masked_maps': False,
'low_pass': None,
'maps_img': '/home/remi-gau/nilearn_data/msdl_atlas/MSDL_rois/msdl_rois.nii',
'mask_img': None,
'reports': True,
'smoothing_fwhm': None,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': 2,
'target_affine': array([[ 4., 0., 0., -78.],
[ 0., 4., 0., -111.],
[ 0., 0., 4., -51.],
[ 0., 0., 0., 1.]]),
'target_shape': (40, 48, 35)}, confounds=[ array([[-0.089762, ..., -0.062316],
...,
[-0.065223, ..., -0.022868]], shape=(168, 5)),
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_desc-reducedConfounds_regressors.tsv'], sample_mask=None, dtype=None, memory=Memory(location=nilearn_cache/joblib), memory_level=1, verbose=1, sklearn_output_config=None)
[NiftiMapsMasker.wrapped] Loading data from
'/home/remi-gau/nilearn_data/development_fmri/development_fmri/sub-pixar003_task-pixar_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz'
[NiftiMapsMasker.wrapped] Resampling images
[NiftiMapsMasker.wrapped] Extracting region signals
[NiftiMapsMasker.wrapped] Cleaning extracted signals
/home/remi-gau/github/nilearn/nilearn/examples/03_connectivity/plot_multi_subject_connectome.py:89: DeprecationWarning: From release 0.14.0, confounds will be standardized using the sample std instead of the population std.
region_ts = masker.fit_transform(
_______________________________________________filter_and_extract - 2.3s, 0.0min
Computing group-sparse precision matrices¶
from nilearn.connectome import GroupSparseCovarianceCV
gsc = GroupSparseCovarianceCV(verbose=1)
gsc.fit(subject_time_series)
from sklearn.covariance import GraphicalLassoCV
gl = GraphicalLassoCV(verbose=True)
gl.fit(np.concatenate(subject_time_series))
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.8s finished
[GroupSparseCovarianceCV.fit] [GroupSparseCovarianceCV] Done refinement 0 out of 4
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 4.8s finished
[GroupSparseCovarianceCV.fit] [GroupSparseCovarianceCV] Done refinement 1 out of 4
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 5.5s finished
[GroupSparseCovarianceCV.fit] [GroupSparseCovarianceCV] Done refinement 2 out of 4
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 4.4s finished
[GroupSparseCovarianceCV.fit] [GroupSparseCovarianceCV] Done refinement 3 out of 4
[GroupSparseCovarianceCV.fit] Final optimization
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.1s finished
[GraphicalLassoCV] Done refinement 1 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.1s finished
[GraphicalLassoCV] Done refinement 2 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.1s finished
[GraphicalLassoCV] Done refinement 3 out of 4: 0s
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 0.1s finished
[GraphicalLassoCV] Done refinement 4 out of 4: 0s
Displaying results¶
atlas_img = msdl_atlas_dataset.maps
atlas_region_coords = plotting.find_probabilistic_atlas_cut_coords(atlas_img)
labels = msdl_atlas_dataset.labels
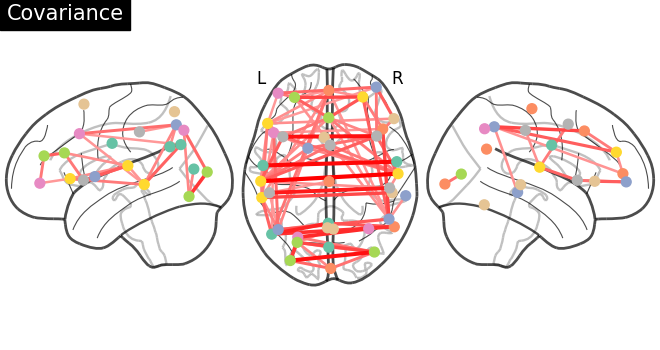
plotting.plot_connectome(
gl.covariance_,
atlas_region_coords,
edge_threshold="90%",
title="Covariance",
display_mode="lzr",
)
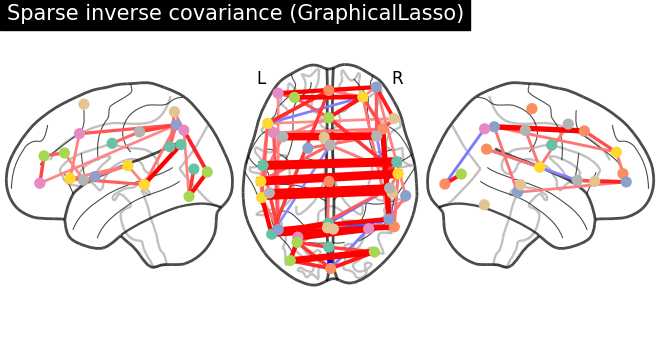
plotting.plot_connectome(
-gl.precision_,
atlas_region_coords,
edge_threshold="90%",
title="Sparse inverse covariance (GraphicalLasso)",
display_mode="lzr",
edge_vmax=0.5,
edge_vmin=-0.5,
)
plot_matrices(gl.covariance_, gl.precision_, "GraphicalLasso", labels)
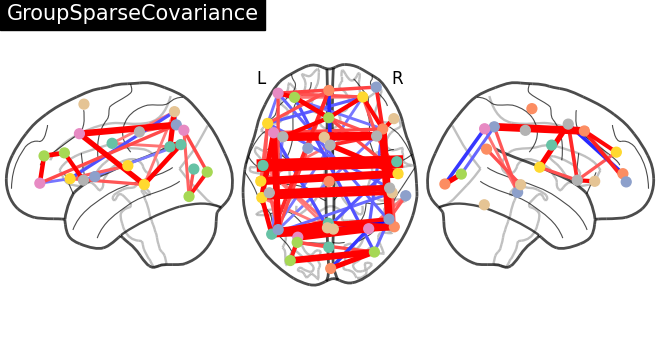
title = "GroupSparseCovariance"
plotting.plot_connectome(
-gsc.precisions_[..., 0],
atlas_region_coords,
edge_threshold="90%",
title=title,
display_mode="lzr",
edge_vmax=0.5,
edge_vmin=-0.5,
)
plot_matrices(gsc.covariances_[..., 0], gsc.precisions_[..., 0], title, labels)
plotting.show()
Total running time of the script: (0 minutes 38.462 seconds)
Estimated memory usage: 1029 MB