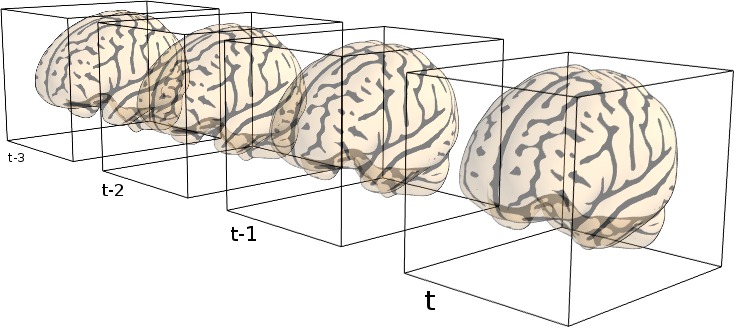
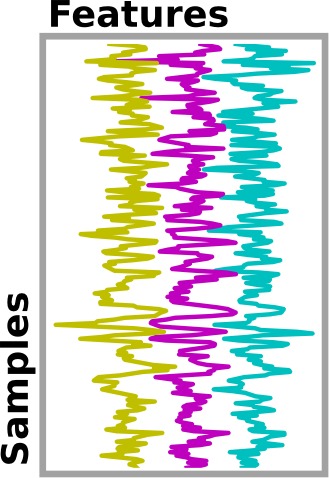
9.3. From neuroimaging volumes to data matrices: the masker objects¶
This chapter introduces the maskers: objects that go from neuroimaging volumes, on the disk or in memory, to data matrices, eg of time series.
9.3.1. The concept of “masker” objects¶
In any analysis, the first step is to load the data. It is often convenient to apply some basic data transformations and to turn the data in a 2D (samples x features) matrix, where the samples could be different time points, and the features derived from different voxels (e.g., restrict analysis to the ventral visual stream), regions of interest (e.g., extract local signals from spheres/cubes), or pre-specified networks (e.g., look at data from all voxels of a set of network nodes). Think of masker objects as swiss-army knives for shaping the raw neuroimaging data in 3D space into the units of observation relevant for the research questions at hand.
Tip
Masker objects can transform both 3D and 4D image objects :
transforming a 3D image produces a 1D (features,) array,
transforming a 4D image produces a 2D (samples, features) array.
“masker” objects (found in module nilearn.maskers)
simplify these “data folding” steps that often precede the
statistical analysis.
Note that the masker objects may not cover all the image transformations
for specific tasks. Users who want to make some specific processing may
have to call specific functions
(modules nilearn.signal, nilearn.masking).
9.3.2. NiftiMasker: applying a mask to load time-series¶
NiftiMasker is a powerful tool to load images and
extract voxel signals in the area defined by the mask.
It applies some basic preprocessing
steps with commonly used parameters as defaults.
But it is very important to look at your data to see the effects
of the preprocessings and validate them.
9.3.2.1. Custom data loading: loading only the first 100 time points¶
Suppose we want to restrict a dataset to the first 100 frames. Below, we load
a movie-watching dataset with fetch_development_fmri(), restrict it to 100 frames and
build a new niimg object that we can give to the masker. Although
possible, there is no need to save your data to a file to pass it to a
NiftiMasker. Simply use nilearn.image.index_img to apply a
slice and create a Niimg in memory:
from nilearn.datasets import fetch_development_fmri
from nilearn.image import index_img
dataset = fetch_development_fmri(n_subjects=1)
epi_filename = dataset.func[0]
epi_img = index_img(epi_filename, slice(0, 100))
9.3.2.2. Controlling how the mask is computed from the data¶
In this section, we show how the masker object can compute a mask automatically for subsequent statistical analysis. On some datasets, the default algorithm may however perform poorly. This is why it is very important to always look at your data before and after feature engineering using masker objects.
Note
The full example described in this section can be found here: plot_mask_computation.py. It is also related to this example: plot_nifti_simple.py.
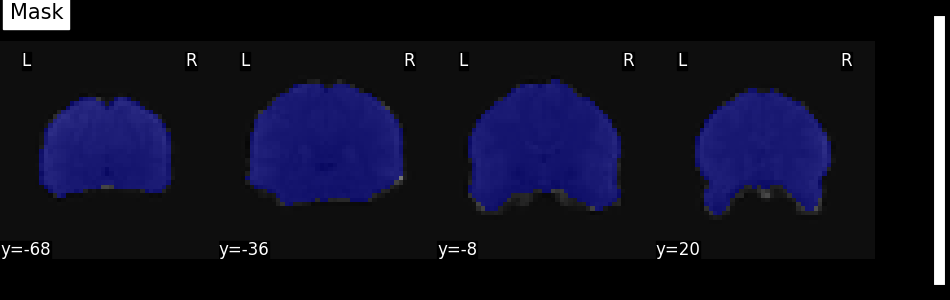
9.3.2.2.1. Visualizing the computed mask¶
If a mask is not specified as an argument, NiftiMasker will try to
compute one from the provided neuroimaging data.
It is very important to verify the quality of the generated mask by visualization.
This allows to see whether it is suitable for your data and intended analyses.
Alternatively, the mask computation parameters can still be modified.
See the NiftiMasker documentation for a complete list of
mask computation parameters.
The mask can be retrieved and visualized from the mask_img_ attribute
of the masker:
from nilearn.image.image import mean_img
from nilearn.plotting import plot_roi, show
mask_img = masker.mask_img_
mean_func_img = mean_img(func_filename)
plot_roi(mask_img, mean_func_img, display_mode="y", cut_coords=4, title="Mask")
show()
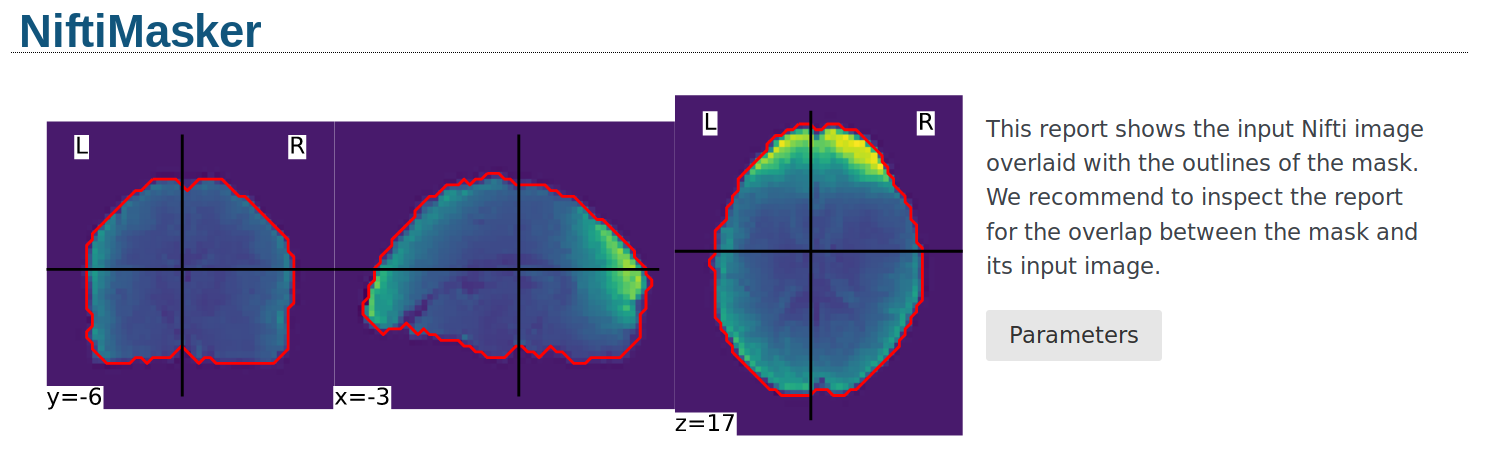
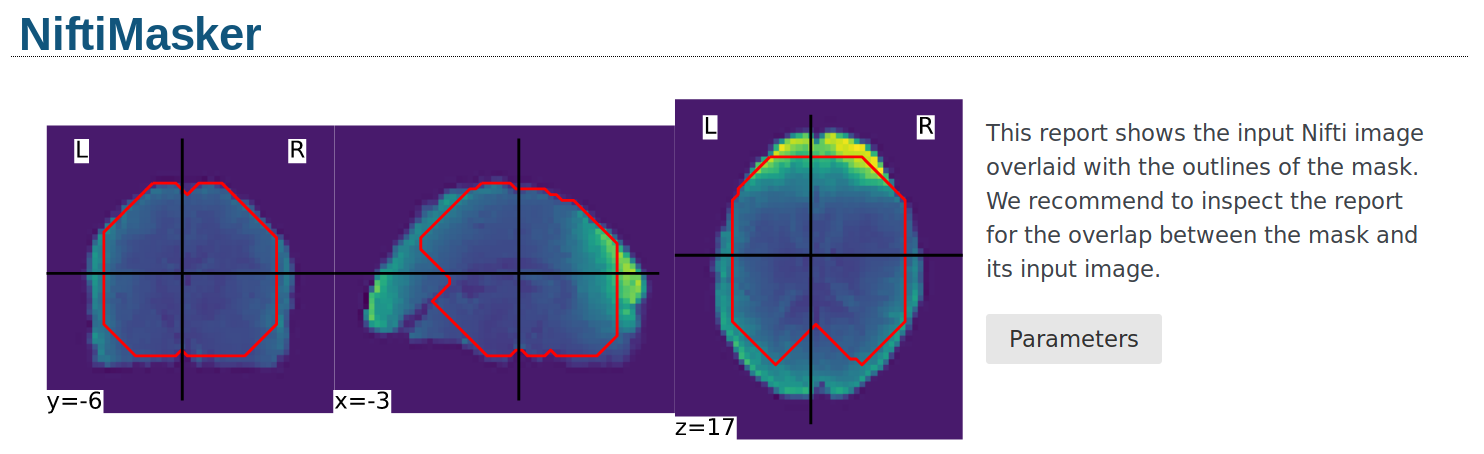
Alternatively, the mask can be visualized using the generate_report
method of the masker. The generated report can be viewed in a Jupyter notebook,
opened in a new browser tab using report.open_in_browser(),
or saved as a portable HTML file report.save_as_html(output_filepath).
9.3.2.2.2. Different masking strategies¶
The mask_strategy argument controls how the mask is computed:
9.3.2.2.3. Extra mask parameters: opening, cutoff…¶
The underlying function is nilearn.masking.compute_epi_mask
called using the mask_args argument of the NiftiMasker.
Controlling these arguments set the fine aspects of the mask. See the
functions documentation, or the NiftiMasker example.
9.3.2.3. Common data preparation steps: smoothing, filtering, resampling¶
NiftiMasker comes with many parameters that enable data
preparation:
>>> import sklearn; sklearn.set_config(print_changed_only=False)
>>> from nilearn import maskers
>>> masker = maskers.NiftiMasker()
>>> masker
NiftiMasker(clean_args=None, cmap='gray', detrend=False, dtype=None, high_pass=None,
high_variance_confounds=False, low_pass=None, mask_args=None,
mask_img=None, mask_strategy='background',
memory=None, memory_level=1, reports=True,
runs=None, smoothing_fwhm=None, standardize=False,
standardize_confounds=True, t_r=None,
target_affine=None, target_shape=None, verbose=0)
The meaning of each parameter is described in the documentation of
NiftiMasker (click on the name NiftiMasker), here we
comment on the most important.
See also
If you do not want to use the NiftiMasker to perform these
simple operations on data, note that they can also be manually
accessed in nilearn such as in
corresponding functions.
9.3.2.3.1. Smoothing¶
NiftiMasker can apply Gaussian spatial smoothing to the
neuroimaging data, useful to fight noise or for inter-individual
differences in neuroanatomy. It is achieved by specifying the
full-width half maximum (FWHM; in millimeter
scale) with the smoothing_fwhm parameter. Anisotropic filtering
is also possible by passing 3 scalars (x, y, z), the
FWHM along the x, y, and z direction.
The underlying function handles properly non-cubic voxels by scaling the given widths appropriately.
See also
9.3.2.3.2. Temporal Filtering and confound removal¶
NiftiMasker can also improve aspects of temporal data
properties, before conversion to voxel signals.
Standardization. Parameter
standardize: Signals can be standardized (scaled to unit variance).Frequency filtering. Low-pass and high-pass filters can be used to remove artifacts. Parameters:
high_passandlow_pass, specified in Hz (note that you must specific the sampling rate in seconds with thet_rparameter:loss_pass=.5, t_r=2.1).Confound removal. Two ways of removing confounds are provided: simple detrending or using prespecified confounds, such as behavioral or movement information.
Linear trends can be removed by activating the
detrendparameter. This accounts for slow (as opposed to abrupt or transient) changes in voxel values along a series of brain images that are unrelated to the signal of interest (e.g., the neural correlates of cognitive tasks). It is not activated by default inNiftiMaskerbut is recommended in almost all scenarios.More complex confounds, measured during the acquision, can be removed by passing them to
NiftiMasker.transform. If the dataset provides a confounds file, just pass its path to the masker. For fMRIPrep outputs, one can useload_confoundsorload_confounds_strategyto select confound variables with some basic sanity check based on fMRIPrep documentation.
Note
Please see the usage example of
load_confounds and
load_confounds_strategy in
plot_signal_extraction.py.
See also
9.3.2.3.3. Resampling: resizing and changing resolutions of images¶
NiftiMasker and many similar classes enable resampling
(recasting of images into different resolutions and transformations of
brain voxel data). Two parameters control resampling:
target_affineto resample (resize, rotate…) images in order to match the spatial configuration defined by the new affine (i.e., matrix transforming from voxel space into world space).Additionally, a
target_shapecan be used to resize images (i.e., cropping or padding with zeros) to match an expected data image dimensions (shape composed of x, y, and z).
How to combine these parameter to obtain the specific resampling desired is explained in details in Resampling images.
9.3.2.4. Inverse transform: unmasking data¶
Note
Inverse transform only performs spatial unmasking.
The data is only brought back into either a 3D or 4D represenetation,
without inverting any signal processing performed by transform.
Once voxel signals have been processed, the result can be visualized as images after unmasking (masked-reduced data transformed back into the original whole-brain space). This step is present in many examples provided in nilearn. Below you will find an excerpt of the example performing Anova-SVM on the Haxby data:
# :class:`~nilearn.plotting.plot_stat_map`
weight_img = decoder.coef_img_["face"]
from nilearn.plotting import plot_stat_map, show
plot_stat_map(weight_img, bg_img=haxby_dataset.anat[0], title="SVM weights")
show()
# %%
Tip
Masker objects can inverse-transform both 1D and 2D arrays :
inverse-transforming a 2D array produces a 4D (X x Y x Z x samples) image,
inverse-transforming a 1D array produces a 3D (X x Y x Z) image.
9.3.3. Extraction of signals from regions: NiftiLabelsMasker, NiftiMapsMasker¶
The purpose of NiftiLabelsMasker and NiftiMapsMasker is to
compute signals from regions containing many voxels. They make it easy to get
these signals once you have an atlas or a parcellation into brain regions.
9.3.3.1. Regions definition¶
Nilearn understands two different ways of defining regions, which are called
labels and maps, handled by NiftiLabelsMasker and
NiftiMapsMasker, respectively.
labels: a single region is defined as the set of all the voxels that have a common label (e.g., anatomical brain region definitions as integers) in the region definition array. The set of regions is defined by a single 3D array, containing a voxel-wise dictionary of label numbers that denote what region a given voxel belongs to. This technique has a big advantage: the required memory load is independent of the number of regions, allowing for a large number of regions. On the other hand, there are several disadvantages: regions cannot spatially overlap and are represented in a binary present/nonpresent coding (no weighting).
maps: a single region is defined as the set of all the voxels that have a non-zero weight. A set of regions is thus defined by a set of 3D images (or a single 4D image), one 3D image per region (as opposed to all regions in a single 3D image such as for labels, cf. above). While these defined weighted regions can exhibit spatial overlap (as opposed to labels), storage cost scales linearly with the number of regions. Handling a large number (e.g., thousands) of regions will prove difficult with this data transformation of whole-brain voxel data into weighted region-wise data.
Note
These usage are illustrated in the section Extracting times series to build a functional connectome.
9.3.3.2. NiftiLabelsMasker Usage¶
Usage of NiftiLabelsMasker is similar to that of
NiftiMapsMasker. The main difference is that it requires a labels image
instead of a set of maps as input.
The background_label keyword of NiftiLabelsMasker deserves
some explanation. The voxels that correspond to the brain or a region
of interest in an fMRI image do not fill the entire image.
Consequently, in the labels image, there must be a label value that corresponds
to “outside” the brain (for which no signal should be extracted).
By default, this label is set to zero in nilearn (referred to as “background”).
Should some non-zero value encoding be necessary, it is possible
to change the background value with the background_label keyword.
9.3.3.3. NiftiMapsMasker Usage¶
This atlas defines its regions using maps. The path to the corresponding
file is given in the maps_img argument.
One important thing that happens transparently during the execution of
NiftiMasker.fit_transform is resampling. Initially, the images
and the atlas do typically not have the same shape nor the same affine.
Casting them into the same format is required for successful signal extraction
The keyword argument resampling_target specifies which format
(i.e., dimensions and affine) the data should be resampled to.
See the reference documentation for NiftiMapsMasker for every
possible option.
9.3.4. Extraction of signals from regions for multiple subjects: MultiNiftiMasker, MultiNiftiLabelsMasker, MultiNiftiMapsMasker¶
The purpose of MultiNiftiMasker, MultiNiftiLabelsMasker and
MultiNiftiMapsMasker is to extend the capabilities of
NiftiMasker, NiftiLabelsMasker and NiftiMapsMasker
as to facilitate the computation of voxel signals in multi-subjects settings.
While NiftiMasker, NiftiLabelsMasker and
NiftiMapsMasker work with 3D inputs (single brain volume) or 4D inputs
(sequence of brain volumes in time for one subject), MultiNiftiMasker,
MultiNiftiLabelsMasker and MultiNiftiMapsMasker
can also handle list of 3D or 4D image objects.
Tip
MultiMasker objects can transform both 3D, 4D, as well as list of 3D or 4D image objects :
transforming a 3D image produces a 1D (features,) array,
transforming a 4D image produces a 2D (samples, features) array,
transforming a list of 3D image produces a list of 1D (features,) array,
transforming a list of 4D image produces a list of 2D (samples, features) array.
9.3.4.1. MultiNiftiMasker Usage¶
MultiNiftiMasker extracts voxel signals for each subject in the areas defined by the
masks.
9.3.4.2. MultiNiftiLabelsMasker Usage¶
MultiNiftiLabelsMasker extracts signals from regions defined by labels
for each subject.
9.3.4.3. MultiNiftiMapsMasker Usage¶
MultiNiftiMapsMasker extracts signals regions defined by maps
for each subject.
9.3.5. Extraction of signals from seeds: NiftiSpheresMasker¶
The purpose of NiftiSpheresMasker is to compute signals from
seeds containing voxels in spheres. It makes it easy to get these signals once
you have a list of coordinates.
A single seed is a sphere defined by the radius (in millimeters) and the
coordinates (typically MNI or TAL) of its center.
Using NiftiSpheresMasker needs to define a list of coordinates.
seeds argument takes a list of 3D coordinates (tuples) of the spheres centers,
they should be in the same space as the images.
Seeds can overlap spatially and are represented in a binary present/nonpresent
coding (no weighting).
Below is an example of a coordinates list of four seeds from the default mode network:
>>> dmn_coords = [(0, -52, 18), (-46, -68, 32), (46, -68, 32), (0, 50, -5)]
radius is an optional argument that takes a real value in millimeters.
If no value is given for the radius argument, the single voxel at the given
seed position is used.
9.3.6. Extraction of signals from surface images SurfaceMasker, SurfaceLabelsMasker, SurfaceMapsMasker, MultiSurfaceMasker¶
The purpose of SurfaceMasker, SurfaceLabelsMasker, SurfaceMapsMasker
is to mirror the capabilities of
NiftiMasker, NiftiLabelsMasker and NiftiMapsMasker
but to extract data from SurfaceImage.
They can perform data extraction from 1D surface data (n_vertices), 2D surface data (n_vertices x samples) or list of 1D or 2D surface data with the same underlying mesh.
Tip
Surface masker objects can transform both 1D, 2D, as well as list of 1D surface image objects.
transforming a 1D image (n_vertices,) produces a 1D array,
transforming a 2D image (n_vertices, samples) produces a 2D array,
transforming a list of
length==nof 1D image (n_vertices,) produces a 2D array (n_vertices, n)
Multi surface masker objects can transform both 1D, 2D, as well as list of 1D or 2D surface image objects.
transforming a 1D image (n_vertices,) produces a 1D array,
transforming a 2D image (n_vertices, samples) produces a 2D array,
transforming a list of
length==nof images (n_vertices,) produces a list oflength==nof arrays where the dimension of each array matches that of the input image
Transforming a 1D image produces a 1D (features,) array. All other input will produce a 1D (samples, features) array..
Surface masker objects can inverse-transform both 1D and 2D arrays :
inverse-transforming a 1D array produces a 1D (n_vertices,) image,
inverse-transforming a 2D array produces a 2D (n_vertices, samples) image.