Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Setting a parameter by cross-validation¶
Here we set the number of features selected in an Anova-SVC approach to maximize the cross-validation score.
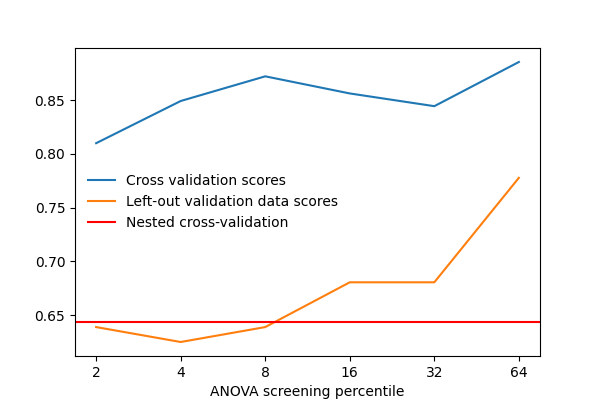
After separating 2 runs for validation, we vary that parameter and measure the cross-validation score. We also measure the prediction score on the left-out validation data. As we can see, the two scores vary by a significant amount: this is due to sampling noise in cross validation, and choosing the parameter k to maximize the cross-validation score, might not maximize the score on left-out data.
Thus using data to maximize a cross-validation score computed on that same data is likely to be too optimistic and lead to an overfit.
The proper approach is known as a “nested cross-validation”. It consists in doing cross-validation loops to set the model parameters inside the cross-validation loop used to judge the prediction performance: the parameters are set separately on each fold, never using the data used to measure performance.
For decoding tasks, in nilearn, this can be done using the
Decoder object, which will automatically select
the best parameters of an estimator from a grid of parameter values.
One difficulty is that the Decoder object is a composite estimator: a pipeline of feature selection followed by Support Vector Machine. Tuning the SVM’s parameters is already done automatically inside the Decoder, but performing cross-validation for the feature selection must be done manually.
Load the Haxby dataset¶
from nilearn import datasets
from nilearn.plotting import show
# by default 2nd subject data will be fetched on which we run our analysis
haxby_dataset = datasets.fetch_haxby()
fmri_img = haxby_dataset.func[0]
mask_img = haxby_dataset.mask
# print basic information on the dataset
print(f"Mask nifti image (3D) is located at: {haxby_dataset.mask}")
print(f"Functional nifti image (4D) are located at: {haxby_dataset.func[0]}")
# Load the behavioral data
import pandas as pd
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
y = labels["labels"]
# Keep only data corresponding to shoes or bottles
from nilearn.image import index_img
condition_mask = y.isin(["shoe", "bottle"])
fmri_niimgs = index_img(fmri_img, condition_mask)
y = y[condition_mask]
run = labels["chunks"][condition_mask]
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
Mask nifti image (3D) is located at: /home/runner/nilearn_data/haxby2001/mask.nii.gz
Functional nifti image (4D) are located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
ANOVA pipeline with Decoder object¶
Nilearn Decoder object aims to provide smooth user experience by acting as a pipeline of several tasks: preprocessing with NiftiMasker, reducing dimension by selecting only relevant features with ANOVA – a classical univariate feature selection based on F-test, and then decoding with different types of estimators (in this example is Support Vector Machine with a linear kernel) on nested cross-validation.
from nilearn.decoding import Decoder
# We provide a grid of hyperparameter values to the Decoder's internal
# cross-validation. If no param_grid is provided, the Decoder will use a
# default grid with sensible values for the chosen estimator
param_grid = [
{
"penalty": ["l2"],
"dual": [True],
"C": [100, 1000],
},
{
"penalty": ["l1"],
"dual": [False],
"C": [100, 1000],
},
]
# Here screening_percentile is set to 2 percent, meaning around 800
# features will be selected with ANOVA.
decoder = Decoder(
estimator="svc",
cv=5,
mask=mask_img,
smoothing_fwhm=4,
standardize="zscore_sample",
screening_percentile=2,
param_grid=param_grid,
verbose=1,
)
Fit the Decoder and predict the responses¶
As a complete pipeline by itself, decoder will perform cross-validation for the estimator, in this case Support Vector Machine. We can output the best parameters selected for each cross-validation fold. See https://scikit-learn.org/stable/modules/cross_validation.html for an excellent explanation of how cross-validation works.
# Fit the Decoder
decoder.fit(fmri_niimgs, y)
# Print the best parameters for each fold
for i, (best_c, best_penalty, best_dual, cv_score) in enumerate(
zip(
decoder.cv_params_["shoe"]["C"],
decoder.cv_params_["shoe"]["penalty"],
decoder.cv_params_["shoe"]["dual"],
decoder.cv_scores_["shoe"],
strict=False,
)
):
print(
f"Fold {i + 1} | Best SVM parameters: C={best_c}"
f", penalty={best_penalty}, dual={best_dual} with score: {cv_score}"
)
# Output the prediction with Decoder
y_pred = decoder.predict(fmri_niimgs)
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453430>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:118: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453430>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 2
[Decoder.fit] Corrected screening-percentile: 1.9171
[Parallel(n_jobs=1)]: Done 5 out of 5 | elapsed: 2.8s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Fold 1 | Best SVM parameters: C=1000, penalty=l1, dual=False with score: 0.9318181818181818
Fold 2 | Best SVM parameters: C=1000, penalty=l2, dual=True with score: 0.9177489177489176
Fold 3 | Best SVM parameters: C=100, penalty=l1, dual=False with score: 0.7705627705627706
Fold 4 | Best SVM parameters: C=1000, penalty=l1, dual=False with score: 0.7727272727272727
Fold 5 | Best SVM parameters: C=1000, penalty=l2, dual=True with score: 0.735930735930736
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453430>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Compute prediction scores with different values of screening percentile¶
import numpy as np
screening_percentile_range = [2, 4, 8, 16, 32, 64]
cv_scores = []
val_scores = []
for sp in screening_percentile_range:
decoder = Decoder(
estimator="svc",
mask=mask_img,
smoothing_fwhm=4,
cv=3,
standardize="zscore_sample",
screening_percentile=sp,
param_grid=param_grid,
verbose=1,
)
decoder.fit(index_img(fmri_niimgs, run < 10), y[run < 10])
cv_scores.append(np.mean(decoder.cv_scores_["bottle"]))
print(f"Sreening Percentile: {sp:.3f}")
print(f"Mean CV score: {cv_scores[-1]:.4f}")
y_pred = decoder.predict(index_img(fmri_niimgs, run == 10))
val_scores.append(np.mean(y_pred == y[run == 10]))
print(f"Validation score: {val_scores[-1]:.4f}")
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7157e0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7157e0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 2
[Decoder.fit] Corrected screening-percentile: 1.9171
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.0s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 2.000
Mean CV score: 0.8167
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c715210>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.5556
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714eb0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714eb0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 4
[Decoder.fit] Corrected screening-percentile: 3.83419
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.4s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 4.000
Mean CV score: 0.8515
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714ee0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.6111
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7144f0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7144f0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 8
[Decoder.fit] Corrected screening-percentile: 7.66838
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 2.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 8.000
Mean CV score: 0.8507
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c715240>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.4444
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7145e0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7145e0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 16
[Decoder.fit] Corrected screening-percentile: 15.3368
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 3.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 16.000
Mean CV score: 0.8681
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c715bd0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.6667
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717be0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717be0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 32
[Decoder.fit] Corrected screening-percentile: 30.6735
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 6.0s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 32.000
Mean CV score: 0.8748
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714730>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.3889
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714370>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:158: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714370>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 64
[Decoder.fit] Corrected screening-percentile: 61.3471
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 10.9s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
Sreening Percentile: 64.000
Mean CV score: 0.8685
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716a10>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Validation score: 0.5000
Nested cross-validation¶
We are going to tune the parameter ‘screening_percentile’ in the pipeline.
import warnings
from sklearn.exceptions import ConvergenceWarning
from sklearn.model_selection import KFold
cv = KFold(n_splits=3)
nested_cv_scores = []
for train, test in cv.split(run):
y_train = np.array(y)[train]
y_test = np.array(y)[test]
val_scores = []
for sp in screening_percentile_range:
with warnings.catch_warnings():
# silence warnings about Liblinear not converging
# increase the number of iterations.
warnings.filterwarnings(
action="ignore", category=ConvergenceWarning
)
decoder = Decoder(
estimator="svc",
mask=mask_img,
smoothing_fwhm=4,
cv=3,
standardize="zscore_sample",
screening_percentile=sp,
param_grid=param_grid,
verbose=1,
)
decoder.fit(index_img(fmri_niimgs, train), y_train)
y_pred = decoder.predict(index_img(fmri_niimgs, test))
val_scores.append(np.mean(y_pred == y_test))
nested_cv_scores.append(np.max(val_scores))
print(f"Nested CV score: {np.mean(nested_cv_scores):.4f}")
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451330>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451330>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 2
[Decoder.fit] Corrected screening-percentile: 1.9171
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14534c0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453ee0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453ee0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 4
[Decoder.fit] Corrected screening-percentile: 3.83419
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1452aa0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14520e0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14520e0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 8
[Decoder.fit] Corrected screening-percentile: 7.66838
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 2.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1450100>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7153f0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7153f0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 16
[Decoder.fit] Corrected screening-percentile: 15.3368
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 3.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451330>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c715930>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c715930>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 32
[Decoder.fit] Corrected screening-percentile: 30.6735
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 4.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14501f0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717e50>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717e50>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 64
[Decoder.fit] Corrected screening-percentile: 61.3471
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 7.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451630>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7164d0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7164d0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 2
[Decoder.fit] Corrected screening-percentile: 1.9171
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1450d60>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716500>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716500>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 4
[Decoder.fit] Corrected screening-percentile: 3.83419
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.0s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14517e0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716590>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716590>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 8
[Decoder.fit] Corrected screening-percentile: 7.66838
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.8s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1450100>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7141f0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7141f0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 16
[Decoder.fit] Corrected screening-percentile: 15.3368
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 2.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451ea0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714910>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714910>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 32
[Decoder.fit] Corrected screening-percentile: 30.6735
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 4.3s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453490>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714160>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714160>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 64
[Decoder.fit] Corrected screening-percentile: 61.3471
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 7.6s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14508b0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7157b0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c7157b0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 2
[Decoder.fit] Corrected screening-percentile: 1.9171
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.7s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453130>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717c10>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c717c10>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 4
[Decoder.fit] Corrected screening-percentile: 3.83419
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.1s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453610>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714b80>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714b80>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 8
[Decoder.fit] Corrected screening-percentile: 7.66838
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 1.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1451e70>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714820>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c714820>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 16
[Decoder.fit] Corrected screening-percentile: 15.3368
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 2.2s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1450d30>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716320>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716320>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 32
[Decoder.fit] Corrected screening-percentile: 30.6735
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 4.0s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1452dd0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
[Decoder.fit] Loading mask from
'/home/runner/nilearn_data/haxby2001/mask.nii.gz'
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716bf0>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:202: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[Decoder.fit] Resampling mask
[Decoder.fit] Finished fit
[Decoder.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c716bf0>
[Decoder.fit] Smoothing images
[Decoder.fit] Extracting region signals
[Decoder.fit] Cleaning extracted signals
[Decoder.fit] Mask volume = 1.96442e+06mm^3 = 1964.42cm^3
[Decoder.fit] Standard brain volume = 1.88299e+06mm^3
[Decoder.fit] Original screening-percentile: 64
[Decoder.fit] Corrected screening-percentile: 61.3471
/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/svm/_base.py:1250: ConvergenceWarning:
Liblinear failed to converge, increase the number of iterations.
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 8.5s finished
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.fit] Computing image from signals
[Decoder.predict] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef1453bb0>
[Decoder.predict] Smoothing images
[Decoder.predict] Extracting region signals
[Decoder.predict] Cleaning extracted signals
Nested CV score: 0.6574
Plot the prediction scores using matplotlib¶
from matplotlib import pyplot as plt
plt.figure(figsize=(6, 4))
plt.plot(cv_scores, label="Cross validation scores")
plt.plot(val_scores, label="Left-out validation data scores")
plt.xticks(
np.arange(len(screening_percentile_range)), screening_percentile_range
)
plt.axis("tight")
plt.xlabel("ANOVA screening percentile")
plt.axhline(
np.mean(nested_cv_scores), label="Nested cross-validation", color="r"
)
plt.legend(loc="best", frameon=False)
show()
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_grid_search.py:229: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (2 minutes 40.163 seconds)
Estimated memory usage: 1010 MB