Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Basic nilearn example: manipulating and looking at data¶
A simple example showing how to load an existing Nifti file and use basic nilearn functionalities.
# Let us use a Nifti file that is shipped with nilearn
from nilearn.datasets import MNI152_FILE_PATH
# Note that the variable MNI152_FILE_PATH is just a path to a Nifti file
print(f"Path to MNI152 template: {MNI152_FILE_PATH!r}")
Path to MNI152 template: PosixPath('/home/runner/work/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/nilearn/datasets/data/mni_icbm152_t1_tal_nlin_sym_09a_converted.nii.gz')
A first step: looking at our data¶
Let’s quickly plot this file:
from nilearn import plotting
plotting.plot_img(MNI152_FILE_PATH)
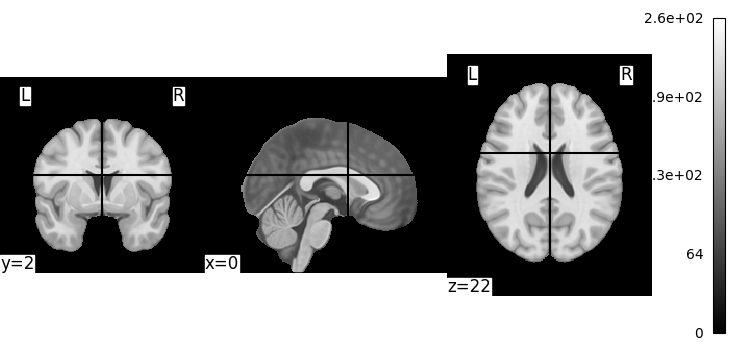
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1f0cd664a0>
This is not a very pretty plot. We just used the simplest possible code. There is a whole section of the documentation on making prettier code.
Exercise: Try plotting one of your own files. In the above, MNI152_FILE_PATH is nothing more than a string with a path pointing to a nifti image. You can replace it with a string pointing to a file on your disk. Note that it should be a 3D volume, and not a 4D volume.
Simple image manipulation: smoothing¶
Let’s use an image-smoothing function from nilearn:
smooth_img
Functions containing ‘img’ can take either a filename or an image as input.
Here we give as inputs the image filename and the smoothing value in mm
from nilearn import image
smooth_anat_img = image.smooth_img(MNI152_FILE_PATH, fwhm=3)
# While we are giving a file name as input, the function returns
# an in-memory object:
smooth_anat_img
<nibabel.nifti1.Nifti1Image object at 0x7f1ee8b106d0>
This is an in-memory object. We can pass it to nilearn function, for instance to look at it
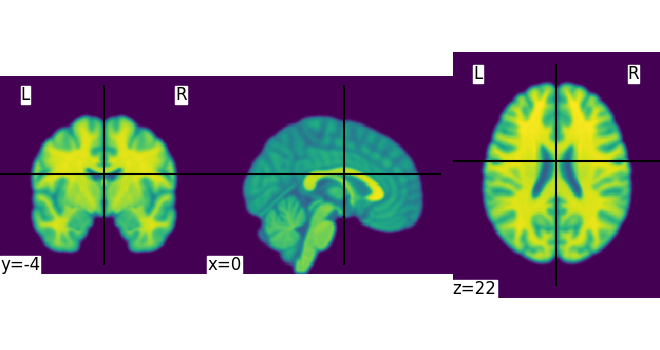
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee8e6fac0>
We could also pass it to the smoothing function
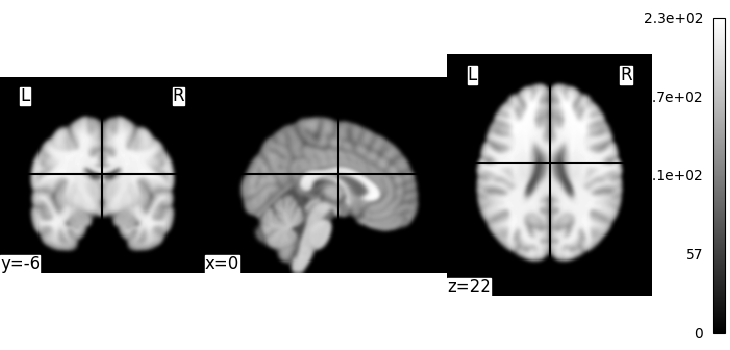
<nilearn.plotting.displays._slicers.OrthoSlicer object at 0x7f1ee830ef80>
Globbing over multiple 3D volumes¶
Nilearn also supports reading multiple volumes at once, using glob-style patterns. For instance, we can smooth volumes from many subjects at once and get a 4D image as output.
First let’s fetch Haxby dataset for subject 1 and 2
from nilearn import datasets
haxby = datasets.fetch_haxby(subjects=[1, 2])
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
[fetch_haxby] Downloading data from
http://data.pymvpa.org/datasets/haxby2001/subj1-2010.01.14.tar.gz ...
[fetch_haxby] Downloaded 26132480 of 314803244 bytes (8.3%%, 11.4s remaining)
[fetch_haxby] Downloaded 75538432 of 314803244 bytes (24.0%%, 6.5s remaining)
[fetch_haxby] Downloaded 124354560 of 314803244 bytes (39.5%%, 4.7s
remaining)
[fetch_haxby] Downloaded 175775744 of 314803244 bytes (55.8%%, 3.2s
remaining)
[fetch_haxby] Downloaded 225902592 of 314803244 bytes (71.8%%, 2.0s
remaining)
[fetch_haxby] Downloaded 275914752 of 314803244 bytes (87.6%%, 0.9s
remaining)
[fetch_haxby] ...done. (7 seconds, 0 min)
[fetch_haxby] Extracting data from
/home/runner/nilearn_data/haxby2001/b2fd65a88d22090da62c3fb828be840e/subj1-2010.
01.14.tar.gz...
[fetch_haxby] .. done.
Now we can find the anatomical images from both subjects using the * wildcard
from pathlib import Path
anats_all_subjects = (
Path(datasets.get_data_dirs()[0]) / "haxby2001" / "subj*" / "anat*"
)
Now we can smooth all the anatomical images at once
This is a 4D image containing one volume per subject
(124, 256, 256, 2)
Saving results to a file¶
We can save any in-memory object as follows:
output_dir = Path.cwd() / "results" / "plot_nilearn_101"
output_dir.mkdir(exist_ok=True, parents=True)
print(f"Output will be saved to: {output_dir}")
anats_all_subjects_smooth.to_filename(
output_dir / "anats_all_subjects_smooth.nii.gz"
)
Output will be saved to: /home/runner/work/nilearn/nilearn/examples/00_tutorials/results/plot_nilearn_101
Finally, calling plotting.show() is necessary to display the figure when running as a script outside IPython
/home/runner/work/nilearn/nilearn/examples/00_tutorials/plot_nilearn_101.py:109: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
To recap, all the nilearn tools can take data as filenames or glob-style patterns or in-memory objects, and return brain volumes as in-memory objects. These can be passed on to other nilearn tools, or saved to disk.
Total running time of the script: (0 minutes 14.374 seconds)
Estimated memory usage: 315 MB