Note
Go to the end to download the full example code or to run this example in your browser via Binder.
A short demo of the surface images & maskers¶
This example shows some more ‘advanced’ features to work with surface images.
This shows:
how to use
SurfaceMaskerand to plotSurfaceImagehow to use
SurfaceLabelsMaskerand to compute a connectome with surface data.how to use run some decoding directly on surface data.
See the dataset description for more information on the data used in this example.
from nilearn._utils.helpers import check_matplotlib
check_matplotlib()
Masking and plotting surface images¶
Here we load the NKI dataset
as a list of SurfaceImage.
Then we extract data with a masker and
compute the mean image across time points for the first subject.
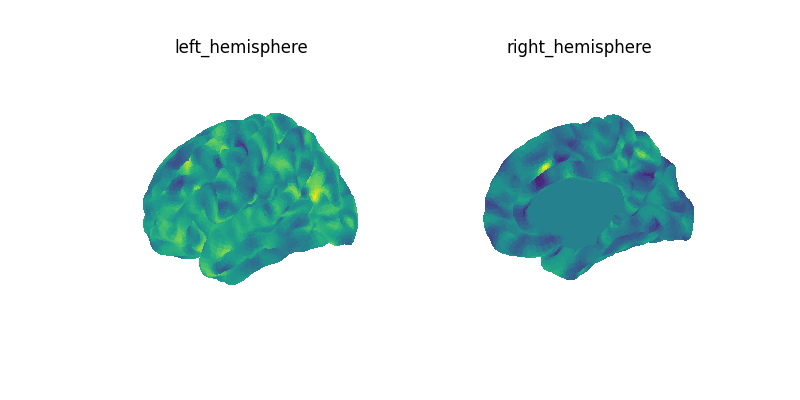
We then plot the the mean image.
import matplotlib.pyplot as plt
import numpy as np
from nilearn.datasets import (
load_fsaverage_data,
load_nki,
)
from nilearn.image import threshold_img
from nilearn.maskers import SurfaceMasker
from nilearn.plotting import plot_matrix, plot_surf, show
surf_img_nki = load_nki()[0]
print(f"NKI image: {surf_img_nki}")
masker = SurfaceMasker(verbose=1)
masked_data = masker.fit_transform(surf_img_nki)
print(f"Masked data shape: {masked_data.shape}")
mean_data = masked_data.mean(axis=0)
mean_img = masker.inverse_transform(mean_data)
print(f"Image mean: {mean_img}")
[load_nki] Dataset found in /home/remi-gau/nilearn_data/nki_enhanced_surface
[load_nki] Loading subject 1 of 1.
NKI image: <SurfaceImage (20484, 895)>
[SurfaceMasker.wrapped] Computing mask
[SurfaceMasker.wrapped] Finished fit
[SurfaceMasker.wrapped] Extracting region signals
[SurfaceMasker.wrapped] Cleaning extracted signals
Masked data shape: (895, 20484)
[SurfaceMasker.inverse_transform] Computing image from signals
Image mean: <SurfaceImage (20484,)>
let’s create a figure with several views for both hemispheres
views = [
"lateral",
"dorsal",
]
hemispheres = ["left", "right", "both"]
for our plots we will be using the fsaverage sulcal data as background map
fsaverage_sulcal = load_fsaverage_data(data_type="sulcal")
mean_img = threshold_img(mean_img, threshold=1e-08, copy=False, two_sided=True)
Let’s ensure that we have the same range centered on 0 for all subplots.
vmax = max(np.absolute(hemi).max() for hemi in mean_img.data.parts.values())
vmin = -vmax
fig, axes = plt.subplots(
nrows=len(views),
ncols=len(hemispheres),
subplot_kw={"projection": "3d"},
figsize=(4 * len(hemispheres), 4),
)
axes = np.atleast_2d(axes)
for view, ax_row in zip(views, axes, strict=False):
for ax, hemi in zip(ax_row, hemispheres, strict=False):
if hemi == "both" and view == "lateral":
view = "left"
elif hemi == "both" and view == "medial":
view = "right"
plot_surf(
surf_map=mean_img,
hemi=hemi,
view=view,
figure=fig,
axes=ax,
title=f"{hemi} - {view}",
colorbar=False,
symmetric_cmap=None,
bg_on_data=True,
vmin=vmin,
vmax=vmax,
bg_map=fsaverage_sulcal,
cmap="seismic",
)
fig.set_size_inches(12, 8)
show()
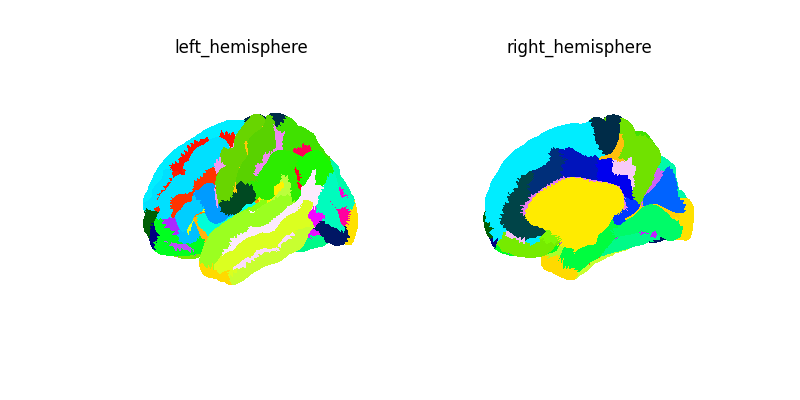
Connectivity with a surface atlas and SurfaceLabelsMasker¶
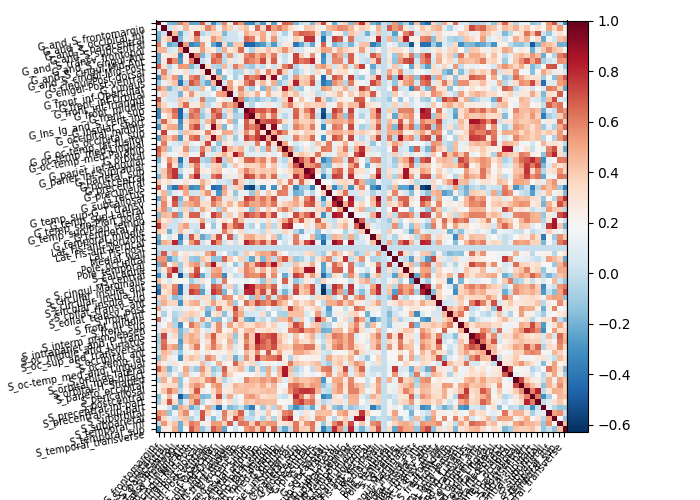
Here we first get the mean time serie for each label of the destrieux atlas for our NKI data. We then compute and plot the connectome of these time series.
from nilearn.connectome import ConnectivityMeasure
from nilearn.datasets import (
fetch_atlas_surf_destrieux,
load_fsaverage,
)
from nilearn.maskers import SurfaceLabelsMasker
from nilearn.surface import SurfaceImage
fsaverage = load_fsaverage("fsaverage5")
destrieux = fetch_atlas_surf_destrieux()
[fetch_atlas_surf_destrieux] Dataset found in /home/remi-gau/nilearn_data/destrieux_surface
/home/remi-gau/github/nilearn/nilearn/examples/07_advanced/plot_surface_image_and_maskers.py:126: UserWarning:
The following regions are present in the atlas look-up table,
but missing from the atlas image:
index name
0 Unknown
destrieux = fetch_atlas_surf_destrieux()
/home/remi-gau/github/nilearn/nilearn/examples/07_advanced/plot_surface_image_and_maskers.py:126: UserWarning:
The following regions are present in the atlas look-up table,
but missing from the atlas image:
index name
0 Unknown
destrieux = fetch_atlas_surf_destrieux()
Let’s create a surface image for this atlas.
labels_img = SurfaceImage(
mesh=fsaverage["pial"],
data={
"left": destrieux["map_left"],
"right": destrieux["map_right"],
},
)
labels_masker = SurfaceLabelsMasker(
labels_img=labels_img, lut=destrieux.lut, verbose=1
).fit()
masked_data = labels_masker.transform(surf_img_nki)
print(f"Masked data shape: {masked_data.shape}")
[SurfaceLabelsMasker.fit] Loading regions from <SurfaceImage (20484,)>
[SurfaceLabelsMasker.fit] Finished fit
[SurfaceLabelsMasker.wrapped] Extracting region signals
[SurfaceLabelsMasker.wrapped] Cleaning extracted signals
Masked data shape: (895, 75)
Plot connectivity matrix¶
connectome_measure = ConnectivityMeasure(
kind="correlation", standardize="zscore_sample"
)
connectome = connectome_measure.fit([masked_data])
vmax = np.absolute(connectome.mean_).max()
vmin = -vmax
We only print every 3rd label for a more legible figure.
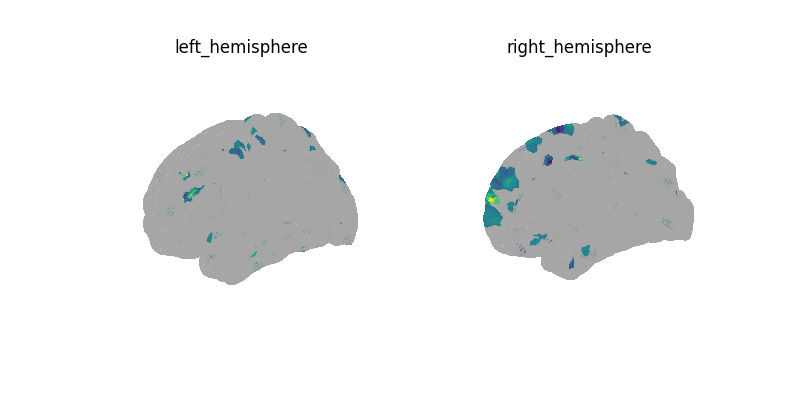
Using the decoder¶
Now using the appropriate masker
we can use a Decoder on surface data
just as we do for volume images.
Note
Here we are given dummy 0 or 1 labels to each time point of the time series. We then decode at each time point. In this sense, the results do not show anything meaningful in a biological sense.
from nilearn.decoding import Decoder
# create some random labels
rng = np.random.RandomState(0)
n_time_points = surf_img_nki.shape[1]
y = rng.choice(
[0, 1],
replace=True,
size=n_time_points,
)
decoder = Decoder(
mask=SurfaceMasker(verbose=1),
param_grid={"C": [0.01, 0.1]},
cv=3,
screening_percentile=1,
)
decoder.fit(surf_img_nki, y)
print("CV scores:", decoder.cv_scores_)
plot_surf(
surf_map=decoder.coef_img_[0],
threshold=1e-6,
bg_map=fsaverage_sulcal,
bg_on_data=True,
cmap="inferno",
vmin=0,
)
show()
/home/remi-gau/github/nilearn/nilearn/examples/07_advanced/plot_surface_image_and_maskers.py:209: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter standardize :
Masker parameter False - overriding estimator parameter True
decoder.fit(surf_img_nki, y)
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:110: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:111: RuntimeWarning: invalid value encountered in divide
f = msb / msw
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:110: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:111: RuntimeWarning: invalid value encountered in divide
f = msb / msw
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:110: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/remi-gau/github/nilearn/nilearn/.tox/doc/lib/python3.10/site-packages/sklearn/feature_selection/_univariate_selection.py:111: RuntimeWarning: invalid value encountered in divide
f = msb / msw
CV scores: {np.int64(0): [0.4991939095387371, 0.5115891053391053, 0.4847132034632034], np.int64(1): [0.4991939095387371, 0.5115891053391053, 0.4847132034632034]}
Decoding with a scikit-learn Pipeline¶
from sklearn import feature_selection, linear_model, pipeline, preprocessing
decoder = pipeline.make_pipeline(
SurfaceMasker(verbose=1),
preprocessing.StandardScaler(),
feature_selection.SelectKBest(
score_func=feature_selection.f_regression, k=500
),
linear_model.Ridge(),
)
decoder.fit(surf_img_nki, y)
coef_img = decoder[:-1].inverse_transform(np.atleast_2d(decoder[-1].coef_))
vmax = max(np.absolute(hemi).max() for hemi in coef_img.data.parts.values())
plot_surf(
surf_map=coef_img,
cmap="RdBu_r",
vmin=-vmax,
vmax=vmax,
threshold=1e-6,
bg_map=fsaverage_sulcal,
bg_on_data=True,
)
show()
[Pipeline.fit] Computing mask
[Pipeline.fit] Finished fit
[Pipeline.fit] Extracting region signals
[Pipeline.fit] Cleaning extracted signals
[Pipeline.inverse_transform] Computing image from signals
Total running time of the script: (0 minutes 23.199 seconds)
Estimated memory usage: 1273 MB