Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Massively univariate analysis of a motor task from the Localizer dataset¶
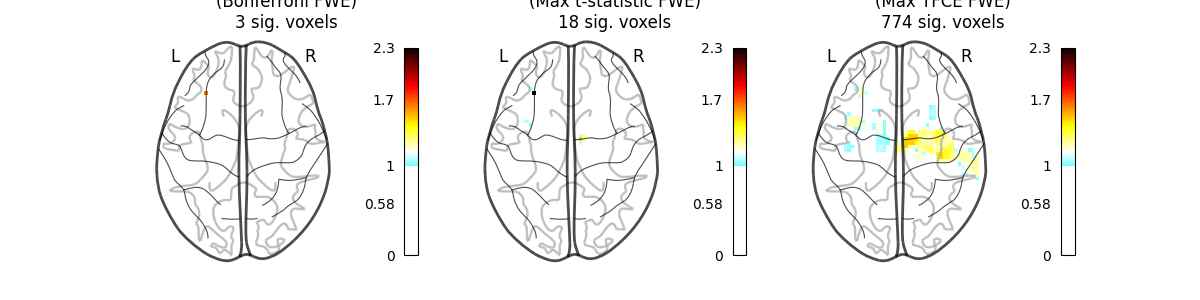
This example shows the results obtained in a massively univariate analysis performed at the inter-subject level with various methods. We use the [left button press (auditory cue)] task from the Localizer dataset and seek association between the contrast values and a variate that measures the speed of pseudo-word reading. No confounding variate is included in the model.
A standard ANOVA is performed. Data smoothed at 5 voxels FWHM are used.
A permuted Ordinary Least Squares algorithm is run at each voxel. Data smoothed at 5 voxels FWHM are used.
Warning
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
from nilearn._utils.helpers import check_matplotlib
check_matplotlib()
import numpy as np
from nilearn import datasets
from nilearn.maskers import NiftiMasker
from nilearn.mass_univariate import permuted_ols
Load Localizer contrast
n_samples = 94
localizer_dataset = datasets.fetch_localizer_contrasts(
["left button press (auditory cue)"],
n_subjects=n_samples,
)
# print basic information on the dataset
print(
"First contrast nifti image (3D) is located "
f"at: {localizer_dataset.cmaps[0]}"
)
tested_var = localizer_dataset.ext_vars["pseudo"]
# Quality check / Remove subjects with bad tested variate
mask_quality_check = np.where(np.logical_not(np.isnan(tested_var)))[0]
n_samples = mask_quality_check.size
contrast_map_filenames = [
localizer_dataset.cmaps[i] for i in mask_quality_check
]
tested_var = tested_var[mask_quality_check].to_numpy().reshape((-1, 1))
print(f"Actual number of subjects after quality check: {int(n_samples)}")
[fetch_localizer_contrasts] Dataset found in
/home/runner/nilearn_data/brainomics_localizer
First contrast nifti image (3D) is located at: /home/runner/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftAuditoryClick.nii.gz
Actual number of subjects after quality check: 89
Mask data
nifti_masker = NiftiMasker(
smoothing_fwhm=5, memory="nilearn_cache", memory_level=1, verbose=1
)
fmri_masked = nifti_masker.fit_transform(contrast_map_filenames)
[NiftiMasker.wrapped] Loading data from
['/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftA
uditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S02/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S03/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S04/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S05/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S06/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S07/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S08/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S09/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S10/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S11/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S12/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S13/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S14/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S16/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S17/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S18/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S19/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S20/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S21/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S22/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S23/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S24/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S25/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S26/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S27/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S28/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S29/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S30/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S31/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S32/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S33/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S34/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S35/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S36/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S37/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S39/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S40/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S41/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S42/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S43/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S44/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S45/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S46/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S47/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S48/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S49/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S50/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S51/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S52/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S53/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S54/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S55/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S56/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S57/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S58/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S59/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S60/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S61/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S63/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S64/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S65/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S66/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S67/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S68/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S69/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S70/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S71/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S72/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S73/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S74/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S75/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S76/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S77/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S78/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S79/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S80/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S82/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S83/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S84/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S85/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S86/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S88/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S89/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S90/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S91/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S92/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S93/cmaps_LeftAu
ditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S94/cmaps_LeftAu
ditoryClick.nii.gz']
[NiftiMasker.wrapped] Computing mask
________________________________________________________________________________
[Memory] Calling nilearn.masking.compute_background_mask...
compute_background_mask([ '/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S02/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S03/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S04/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S05/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S06/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S07/cmaps_LeftAu..., verbose=0)
__________________________________________compute_background_mask - 0.6s, 0.0min
[NiftiMasker.wrapped] Resampling mask
________________________________________________________________________________
[Memory] Calling nilearn.image.resampling.resample_img...
resample_img(<nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>, target_affine=None, target_shape=None, copy=False, interpolation='nearest')
_____________________________________________________resample_img - 0.0s, 0.0min
[NiftiMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.maskers.nifti_masker.filter_and_mask...
filter_and_mask([ '/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S02/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S03/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S04/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S05/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S06/cmaps_LeftAuditoryClick.nii.gz',
'/home/runner/nilearn_data/brainomics_localizer/brainomics_data/S07/cmaps_LeftAu...,
<nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>, { 'clean_args': None,
'clean_kwargs': {},
'cmap': 'gray',
'detrend': False,
'dtype': None,
'high_pass': None,
'high_variance_confounds': False,
'low_pass': None,
'reports': True,
'runs': None,
'smoothing_fwhm': 5,
'standardize': False,
'standardize_confounds': True,
't_r': None,
'target_affine': None,
'target_shape': None}, memory_level=1, memory=Memory(location=nilearn_cache/joblib), verbose=1, confounds=None, sample_mask=None, copy=True, dtype=None, sklearn_output_config=None)
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ebf3be230>
[NiftiMasker.wrapped] Smoothing images
[NiftiMasker.wrapped] Extracting region signals
[NiftiMasker.wrapped] Cleaning extracted signals
__________________________________________________filter_and_mask - 1.3s, 0.0min
Anova (parametric F-scores)
from sklearn.feature_selection import f_regression
_, pvals_anova = f_regression(fmri_masked, tested_var.ravel(), center=True)
pvals_anova *= fmri_masked.shape[1]
pvals_anova[np.isnan(pvals_anova)] = 1
pvals_anova[pvals_anova > 1] = 1
neg_log_pvals_anova = -np.log10(pvals_anova)
neg_log_pvals_anova_unmasked = nifti_masker.inverse_transform(
neg_log_pvals_anova
)
[NiftiMasker.inverse_transform] Computing image from signals
________________________________________________________________________________
[Memory] Calling nilearn.masking.unmask...
unmask(array([-0., ..., -0.], shape=(41852,)), <nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>)
___________________________________________________________unmask - 0.2s, 0.0min
Perform massively univariate analysis with permuted OLS
This method will produce both voxel-level FWE-corrected -log10 p-values and TFCE-based FWE-corrected -log10 p-values.
Note
permuted_ols can support a wide range
of analysis designs, depending on the tested_var.
For example, if you wished to perform a one-sample test, you could
simply provide an array of ones (e.g., np.ones(n_samples)).
ols_outputs = permuted_ols(
tested_var, # this is equivalent to the design matrix, in array form
fmri_masked,
model_intercept=True,
masker=nifti_masker,
tfce=True,
n_perm=100, # 100 for the sake of time. Ideally, this should be 10000.
verbose=1, # display progress bar
n_jobs=2, # can be changed to use more CPUs
)
neg_log_pvals_permuted_ols_unmasked = nifti_masker.inverse_transform(
ols_outputs["logp_max_t"][0, :] # select first regressor
)
neg_log_pvals_tfce_unmasked = nifti_masker.inverse_transform(
ols_outputs["logp_max_tfce"][0, :] # select first regressor
)
[NiftiMasker.inverse_transform] Computing image from signals
________________________________________________________________________________
[Memory] Calling nilearn.masking.unmask...
unmask(array([[ 1.604273, ..., -0.864518]], shape=(1, 41852)), <nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>)
___________________________________________________________unmask - 0.2s, 0.0min
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 2 out of 2 | elapsed: 25.1s finished
[NiftiMasker.inverse_transform] Computing image from signals
________________________________________________________________________________
[Memory] Calling nilearn.masking.unmask...
unmask(array([-0., ..., -0.], shape=(41852,)), <nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>)
___________________________________________________________unmask - 0.2s, 0.0min
[NiftiMasker.inverse_transform] Computing image from signals
________________________________________________________________________________
[Memory] Calling nilearn.masking.unmask...
unmask(array([ 0.031194, ..., -0. ], shape=(41852,)), <nibabel.nifti1.Nifti1Image object at 0x7f1ee8771120>)
___________________________________________________________unmask - 0.2s, 0.0min
Visualization
import matplotlib.pyplot as plt
from nilearn import plotting
from nilearn.image import get_data
threshold = -np.log10(0.1) # 10% corrected
vmax = max(
np.amax(ols_outputs["logp_max_t"]),
np.amax(neg_log_pvals_anova),
np.amax(ols_outputs["logp_max_tfce"]),
)
images_to_plot = {
"Parametric Test\n(Bonferroni FWE)": neg_log_pvals_anova_unmasked,
"Permutation Test\n(Max t-statistic FWE)": (
neg_log_pvals_permuted_ols_unmasked
),
"Permutation Test\n(Max TFCE FWE)": neg_log_pvals_tfce_unmasked,
}
fig, axes = plt.subplots(figsize=(10, 4), ncols=3)
for i_col, (title, img) in enumerate(images_to_plot.items()):
ax = axes[i_col]
n_detections = (get_data(img) > threshold).sum()
new_title = f"{title}\n{n_detections} sig. voxels"
plotting.plot_glass_brain(
img,
vmax=vmax,
display_mode="z",
threshold=threshold,
vmin=threshold,
cmap="inferno",
figure=fig,
axes=ax,
)
ax.set_title(new_title)
fig.suptitle(
"Group left button press ($-\\log_{10}$ p-values)",
y=1,
fontsize=16,
)
fig.subplots_adjust(top=0.75, wspace=0.5)
plotting.show()
/home/runner/work/nilearn/nilearn/examples/07_advanced/plot_localizer_mass_univariate_methods.py:156: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (0 minutes 35.909 seconds)
Estimated memory usage: 212 MB