Note
Go to the end to download the full example code or to run this example in your browser via Binder
A short demo of the surface images & maskers#
copied from the nilearn sandbox discussion, to be transformed into tests & examples
Note
this example is meant to support discussion around a tentative API for surface images in nilearn. This functionality is provided by the nilearn.experimental.surface module; it is still incomplete and subject to change without a deprecation cycle. Please participate in the discussion on GitHub!
try:
import matplotlib.pyplot as plt
except ImportError:
raise RuntimeError("This script needs the matplotlib library")
from typing import Optional, Sequence
from nilearn import plotting
from nilearn.experimental import surface
def plot_surf_img(
img: surface.SurfaceImage,
parts: Optional[Sequence[str]] = None,
mesh: Optional[surface.PolyMesh] = None,
**kwargs,
) -> plt.Figure:
if mesh is None:
mesh = img.mesh
if parts is None:
parts = list(img.data.keys())
fig, axes = plt.subplots(
1,
len(parts),
subplot_kw={"projection": "3d"},
figsize=(4 * len(parts), 4),
)
for ax, mesh_part in zip(axes, parts):
plotting.plot_surf(
mesh[mesh_part],
img.data[mesh_part],
axes=ax,
title=mesh_part,
**kwargs,
)
assert isinstance(fig, plt.Figure)
return fig
img = surface.fetch_nki()[0]
print(f"NKI image: {img}")
masker = surface.SurfaceMasker()
masked_data = masker.fit_transform(img)
print(f"Masked data shape: {masked_data.shape}")
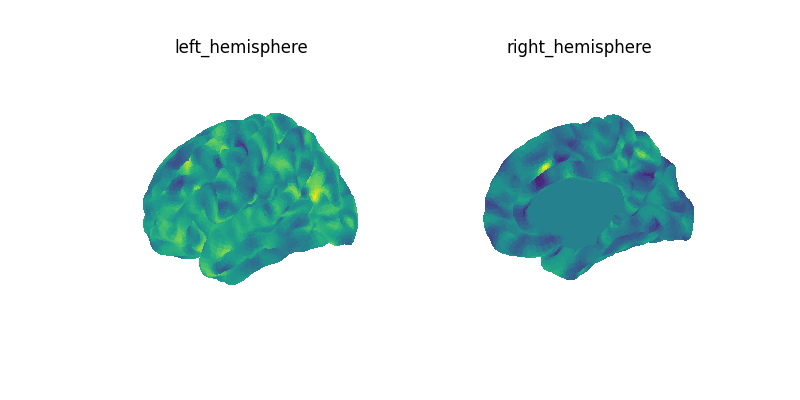
mean_data = masked_data.mean(axis=0)
mean_img = masker.inverse_transform(mean_data)
print(f"Image mean: {mean_img}")
plot_surf_img(mean_img)
plotting.show()
NKI image: <SurfaceImage (895, 20484)>
Masked data shape: (895, 20484)
Image mean: <SurfaceImage (20484,)>
Connectivity with a surface atlas and SurfaceLabelsMasker#
from nilearn import connectome, plotting
img = surface.fetch_nki()[0]
print(f"NKI image: {img}")
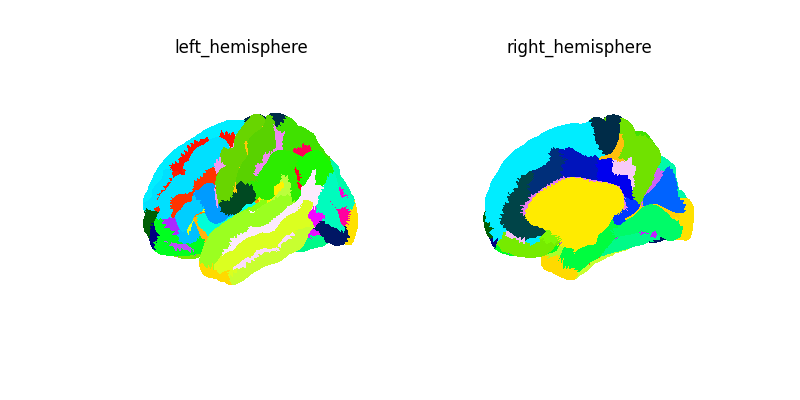
labels_img, label_names = surface.fetch_destrieux()
print(f"Destrieux image: {labels_img}")
plot_surf_img(labels_img, cmap="gist_ncar", avg_method="median")
labels_masker = surface.SurfaceLabelsMasker(labels_img, label_names).fit()
masked_data = labels_masker.transform(img)
print(f"Masked data shape: {masked_data.shape}")
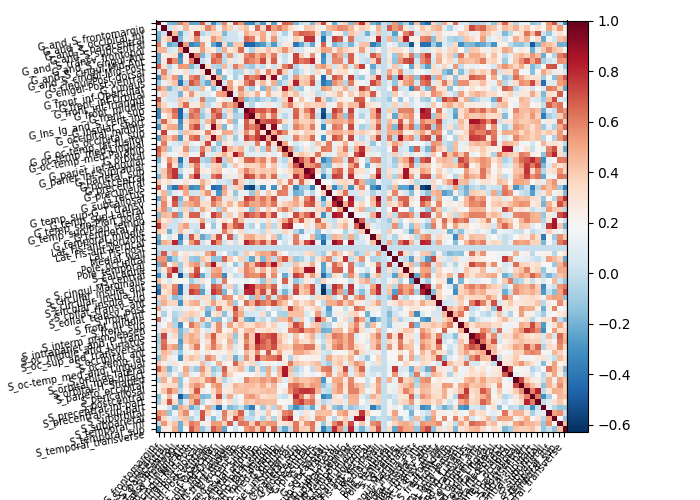
connectome = (
connectome.ConnectivityMeasure(kind="correlation").fit([masked_data]).mean_
)
plotting.plot_matrix(connectome, labels=labels_masker.label_names_)
plotting.show()
NKI image: <SurfaceImage (895, 20484)>
Destrieux image: <SurfaceImage (20484,)>
Masked data shape: (895, 75)
Using the Decoder#
import numpy as np
from nilearn import decoding, plotting
from nilearn._utils import param_validation
The following is just disabling a couple of checks performed by the decoder that would force us to use a NiftiMasker.
def monkeypatch_masker_checks():
def adjust_screening_percentile(screening_percentile, *args, **kwargs):
return screening_percentile
param_validation.adjust_screening_percentile = adjust_screening_percentile
monkeypatch_masker_checks()
Now using the appropriate masker we can use a Decoder on surface data just as we do for volume images.
img = surface.fetch_nki()[0]
y = np.random.RandomState(0).choice([0, 1], replace=True, size=img.shape[0])
decoder = decoding.Decoder(
mask=surface.SurfaceMasker(),
param_grid={"C": [0.01, 0.1]},
cv=3,
screening_percentile=1,
)
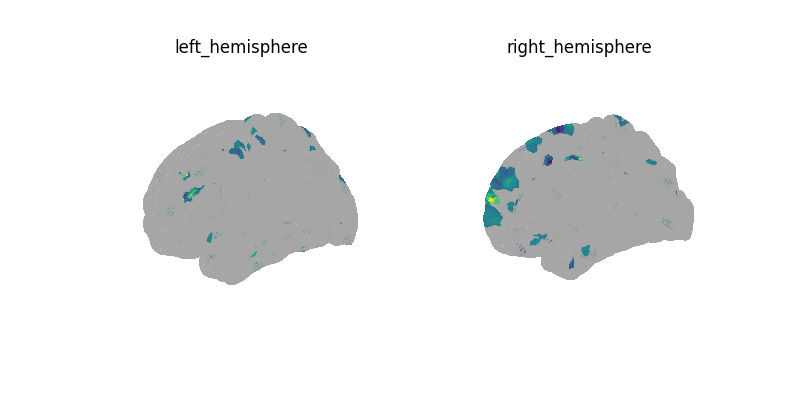
decoder.fit(img, y)
print("CV scores:", decoder.cv_scores_)
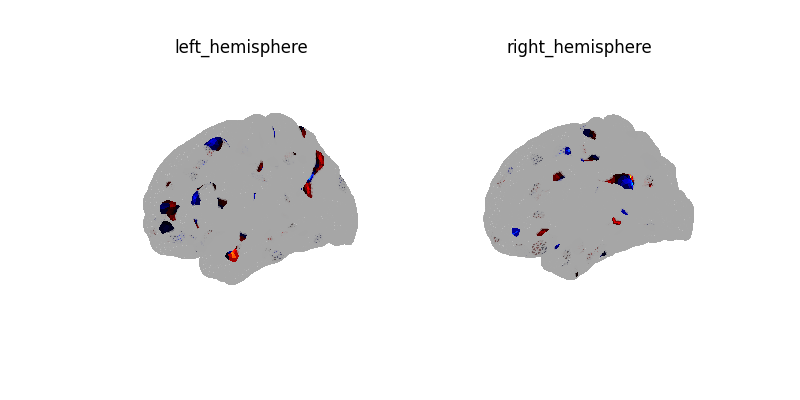
plot_surf_img(decoder.coef_img_[0], threshold=1e-6)
plotting.show()
/home/himanshu/Desktop/nilearn_work/nilearn/nilearn/_utils/masker_validation.py:113: UserWarning: Overriding provided-default estimator parameters with provided masker parameters :
Parameter standardize :
Masker parameter False - overriding estimator parameter True
warnings.warn(warn_str)
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning: invalid value encountered in divide
f = msb / msw
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning: invalid value encountered in divide
f = msb / msw
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:112: UserWarning: Features [ 8 36 38 ... 20206 20207 20208] are constant.
warnings.warn("Features %s are constant." % constant_features_idx, UserWarning)
/home/himanshu/.local/miniconda3/envs/nilearnpy/lib/python3.12/site-packages/sklearn/feature_selection/_univariate_selection.py:113: RuntimeWarning: invalid value encountered in divide
f = msb / msw
CV scores: {0: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034], 1: [0.4991939095387371, 0.5115891053391053, 0.4847132034632034]}
Decoding with a scikit-learn Pipeline#
import numpy as np
from sklearn import feature_selection, linear_model, pipeline, preprocessing
from nilearn import plotting
img = surface.fetch_nki()[0]
y = np.random.RandomState(0).normal(size=img.shape[0])
decoder = pipeline.make_pipeline(
surface.SurfaceMasker(),
preprocessing.StandardScaler(),
feature_selection.SelectKBest(
score_func=feature_selection.f_regression, k=500
),
linear_model.Ridge(),
)
decoder.fit(img, y)
coef_img = decoder[:-1].inverse_transform(np.atleast_2d(decoder[-1].coef_))
vmax = max([np.absolute(dp).max() for dp in coef_img.data.values()])
plot_surf_img(
coef_img,
cmap="cold_hot",
vmin=-vmax,
vmax=vmax,
threshold=1e-6,
)
plotting.show()
Total running time of the script: (0 minutes 33.525 seconds)
Estimated memory usage: 1532 MB