Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Example of generic design in second-level models¶
This example shows the results obtained in a group analysis using a more complex contrast than a one- or two-sample t test. We use the [left button press (auditory cue)] task from the Localizer dataset and seek association between the contrast values and a variate that measures the speed of pseudo-word reading. No confounding variate is included in the model.
At first, we need to load the Localizer contrasts.
from nilearn.datasets import fetch_localizer_contrasts
n_samples = 94
localizer_dataset = fetch_localizer_contrasts(
["left button press (auditory cue)"],
n_subjects=n_samples,
)
[fetch_localizer_contrasts] Dataset found in
/home/runner/nilearn_data/brainomics_localizer
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d27da3a114a4200190453ab/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d27ebc3114a42001704a18d/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d27f1f0114a42001804603e/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28092e45253a001c3e597f/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d281a531c5b4a001c9ea662/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28295aa26b340018087ef4/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d283473a26b34001609ed88/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d284374114a42001605f4d2/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28590d114a4200160607da/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d285a53114a4200160608be/ ...
[fetch_localizer_contrasts] ...done. (32 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d286f35a26b34001908e5c1/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2888ce1c5b4a001b9f789c/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d289b2945253a00193d32ac/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28a00245253a001c3efac9/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28bc0145253a00193d53ab/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28cfd91c5b4a001c9f404d/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28db3ba26b34001808f444/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28f0bc1c5b4a001b9fd7f3/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d28ffc245253a00193d7dac/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2909cd1c5b4a001b9fe6c5/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2919e2114a42001606b46c/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2928bc45253a001b3cf010/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d293a50a26b34001909682a/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d29492fa26b34001709070f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d295328114a42001606dd9a/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2c37031c5b4a001ca0da2b/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2c442e114a420017071134/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2c5c431c5b4a001da257a5/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2c6c2645253a001c42460f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2ec286251f0e001604a189/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2ed2875d2cdc001702b4c5/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d341711a667db0017fc816f/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d34294d835aff001958add9/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2ef8925d2cdc001702e0a5/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f0851251f0e0018044fe4/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f26e4a667db0017f72ae9/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f358c251f0e001704a76a/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f41d2835aff001a52da0c/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f5acc835aff0018532004/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f692d835aff00175372e9/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f7456835aff0017537992/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f8881a667db0018f6b634/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2f9552251f0e001605bb64/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2faf785d2cdc0017039bb1/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2fbffd835aff0018535ef5/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2fc225a667db001af6222a/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2fdd77835aff00195494d4/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2fe5d5a667db0017f80f32/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d2ff3ea835aff0018538140/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d301049a667db0019f67ca0/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3021b65d2cdc00190344d6/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d302afe5d2cdc0018030034/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d303ad4835aff001853bca4/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d304f845d2cdc001a032801/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3058cd835aff001853d4c7/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d306f15a667db0018f78b4d/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d307f8b251f0e00190519ca/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d309cb5251f0e001606fe4b/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d30a667251f0e00190534dc/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d30bb07251f0e001705df42/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d30df37251f0e001705fd72/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d30e232a667db0018f7f2a9/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d30f7ec251f0e001805e3cd/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3116dca667db0018f81c29/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d312688251f0e0016079f29/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3134fe5d2cdc001705393d/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3143f9835aff00195630ce/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d315ac0835aff001754e139/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3160bc835aff00195649cf/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d317bb2251f0e001608002e/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d318a6c251f0e001905b6ed/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d31962da667db0017fa303d/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d31abe45d2cdc0019046202/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d31cdeea667db001af75ab9/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d31dc83835aff001956e6c5/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d31fefea667db0018f8ea9f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32072b5d2cdc0018043969/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3219b4a667db0018f8ff1f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3234f3835aff00175590f0/ ...
[fetch_localizer_contrasts] ...done. (3 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d323d06251f0e001706f0be/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d324e77251f0e001806e5e7/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d326b70835aff00195762f8/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32731a5d2cdc001a0472bb/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3286b7251f0e001906427f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3298815d2cdc001804700c/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32ab6ea667db0017fb59e8/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32bb275d2cdc001a049841/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32d901a667db0018f9684f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32e9d3835aff001957cd79/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d32f34f835aff001755ee97/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d3306db5d2cdc001706c36f/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d332373251f0e001609acbd/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
[fetch_localizer_contrasts] Downloading data from
https://osf.io/download/5d332b7e835aff001957feec/ ...
[fetch_localizer_contrasts] ...done. (2 seconds, 0 min)
Let’s print basic information on the dataset.
print(
"First contrast nifti image (3D) is located "
f"at: {localizer_dataset.cmaps[0]}"
)
First contrast nifti image (3D) is located at: /home/runner/nilearn_data/brainomics_localizer/brainomics_data/S01/cmaps_LeftAuditoryClick.nii.gz
we also need to load the behavioral variable.
tested_var = localizer_dataset.ext_vars["pseudo"]
print(tested_var)
0 15.0
1 16.0
2 14.0
3 19.0
4 16.0
...
89 12.0
90 16.0
91 13.0
92 25.0
93 21.0
Name: pseudo, Length: 94, dtype: float64
It is worth to do a quality check and remove subjects with missing values.
import numpy as np
mask_quality_check = np.where(np.logical_not(np.isnan(tested_var)))[0]
n_samples = mask_quality_check.size
contrast_map_filenames = [
localizer_dataset.cmaps[i] for i in mask_quality_check
]
tested_var = tested_var[mask_quality_check].to_numpy().reshape((-1, 1))
print(f"Actual number of subjects after quality check: {int(n_samples)}")
Actual number of subjects after quality check: 89
Estimate second level model¶
We define the input maps and the design matrix for the second level model and fit it.
import pandas as pd
design_matrix = pd.DataFrame(
np.hstack((tested_var, np.ones_like(tested_var))),
columns=["fluency", "intercept"],
)
Fit of the second-level model
from nilearn.glm.second_level import SecondLevelModel
model = SecondLevelModel(smoothing_fwhm=5.0, n_jobs=2, verbose=1)
model.fit(contrast_map_filenames, design_matrix=design_matrix)
[SecondLevelModel.fit] Fitting second level model. Take a deep breath.
[SecondLevelModel.fit] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1ef14d67d0>
[SecondLevelModel.fit] Computing mask
[SecondLevelModel.fit] Resampling mask
[SecondLevelModel.fit] Finished fit
[SecondLevelModel.fit]
Computation of second level model done in 0.32 seconds.
To estimate the contrast is very simple. We can just provide the column name of the design matrix.
z_map = model.compute_contrast("fluency", output_type="z_score")
[SecondLevelModel.compute_contrast] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f1ef14d6bf0>
[SecondLevelModel.compute_contrast] Smoothing images
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
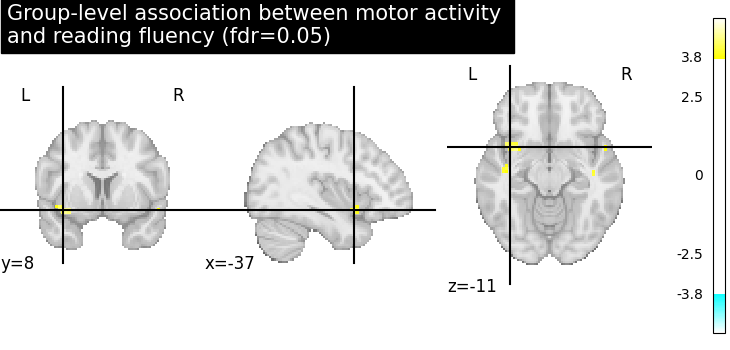
We compute the fdr-corrected p = 0.05 threshold for these data.
from nilearn.glm import threshold_stats_img
_, threshold = threshold_stats_img(z_map, alpha=0.05, height_control="fdr")
Let us plot the second level contrast at the computed thresholds.
from nilearn.plotting import plot_stat_map, show
cut_coords = [10, -5, 10]
plot_stat_map(
z_map,
threshold=threshold,
title="Group-level association between motor activity \n"
"and reading fluency (fdr=0.05)",
cut_coords=cut_coords,
draw_cross=False,
)
show()
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_second_level_association_test.py:92: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Computing the (corrected) p-values with parametric test to compare with non parametric test
from nilearn.image import get_data, math_img
p_val = model.compute_contrast("fluency", output_type="p_value")
n_voxels = np.sum(get_data(model.masker_.mask_img_))
# Correcting the p-values for multiple testing and taking negative logarithm
neg_log_pval = math_img(
f"-np.log10(np.minimum(1, img * {n_voxels!s}))", img=p_val
)
[SecondLevelModel.compute_contrast] Loading data from
<nibabel.nifti1.Nifti1Image object at 0x7f1ece5cdbd0>
[SecondLevelModel.compute_contrast] Smoothing images
[SecondLevelModel.compute_contrast] Extracting region signals
[SecondLevelModel.compute_contrast] Cleaning extracted signals
[SecondLevelModel.compute_contrast] Computing image from signals
<string>:1: RuntimeWarning:
divide by zero encountered in log10
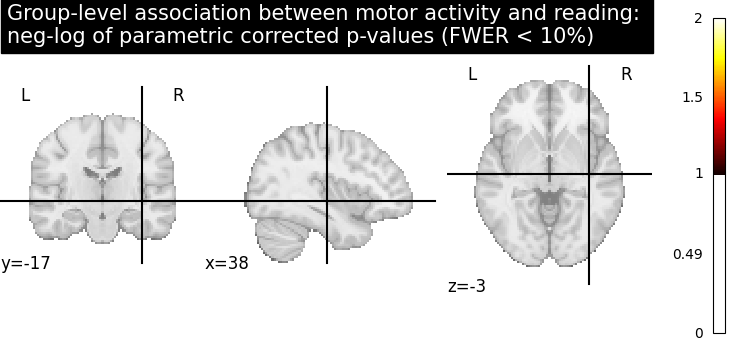
Let us plot the (corrected) negative log p-values for the parametric test
# Since we are plotting negative log p-values and using a threshold equal to 1,
# it corresponds to corrected p-values lower than 10%, meaning that there
# is less than 10% probability to make a single false discovery
# (90% chance that we make no false discoveries at all).
# This threshold is much more conservative than the previous one.
threshold = 1
title = (
"Group-level association between motor activity and reading: \n"
"neg-log of parametric corrected p-values (FWER < 10%)"
)
plot_stat_map(
neg_log_pval,
cut_coords=cut_coords,
threshold=threshold,
title=title,
vmin=threshold,
cmap="inferno",
draw_cross=False,
)
show()
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_second_level_association_test.py:119: UserWarning:
Non-finite values detected. These values will be replaced with zeros.
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_second_level_association_test.py:128: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
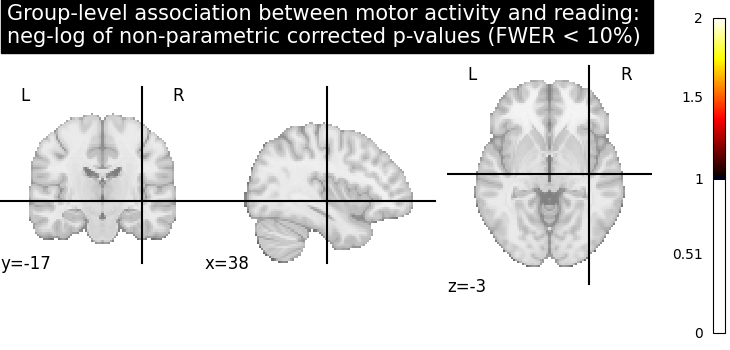
Computing the (corrected) negative log p-values with permutation test
from nilearn.glm.second_level import non_parametric_inference
neg_log_pvals_permuted_ols_unmasked = non_parametric_inference(
contrast_map_filenames,
design_matrix=design_matrix,
second_level_contrast="fluency",
model_intercept=True,
n_perm=1000,
two_sided_test=False,
mask=None,
smoothing_fwhm=5.0,
n_jobs=2,
verbose=1,
)
[non_parametric_inference] Fitting second level model...
[non_parametric_inference]
Computation of second level model done in 0.30462145805358887 seconds
Let us plot the (corrected) negative log p-values
title = (
"Group-level association between motor activity and reading: \n"
"neg-log of non-parametric corrected p-values (FWER < 10%)"
)
plot_stat_map(
neg_log_pvals_permuted_ols_unmasked,
cut_coords=cut_coords,
threshold=threshold,
title=title,
vmin=threshold,
cmap="inferno",
draw_cross=False,
)
show()
# The neg-log p-values obtained with non parametric testing are capped at 3
# since the number of permutations is 1e3.
# The non parametric test yields a few more discoveries
# and is then more powerful than the usual parametric procedure.
/home/runner/work/nilearn/nilearn/examples/05_glm_second_level/plot_second_level_association_test.py:162: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (4 minutes 35.354 seconds)
Estimated memory usage: 241 MB