Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Searchlight analysis of face vs house recognition¶
Searchlight analysis requires fitting a classifier a large amount of times. As a result, it is an intrinsically slow method. In order to speed up computing, in this example, Searchlight is run only on one slice on the fMRI (see the generated figures).
Warning
If you are using Nilearn with a version older than 0.9.0,
then you should either upgrade your version or import maskers
from the input_data module instead of the maskers module.
That is, you should manually replace in the following example all occurrences of:
from nilearn.maskers import NiftiMasker
with:
from nilearn.input_data import NiftiMasker
Load Haxby dataset¶
import pandas as pd
from nilearn.datasets import fetch_haxby
from nilearn.image import get_data, load_img, new_img_like
# We fetch 2nd subject from haxby datasets (which is default)
haxby_dataset = fetch_haxby()
# print basic information on the dataset
print(f"Anatomical nifti image (3D) is located at: {haxby_dataset.mask}")
print(f"Functional nifti image (4D) is located at: {haxby_dataset.func[0]}")
fmri_filename = haxby_dataset.func[0]
labels = pd.read_csv(haxby_dataset.session_target[0], sep=" ")
y = labels["labels"]
run = labels["chunks"]
[fetch_haxby] Dataset found in /home/runner/nilearn_data/haxby2001
Anatomical nifti image (3D) is located at: /home/runner/nilearn_data/haxby2001/mask.nii.gz
Functional nifti image (4D) is located at: /home/runner/nilearn_data/haxby2001/subj2/bold.nii.gz
Restrict to faces and houses¶
from nilearn.image import index_img
condition_mask = y.isin(["face", "house"])
fmri_img = index_img(fmri_filename, condition_mask)
y, run = y[condition_mask], run[condition_mask]
Prepare masks¶
mask_img is the original mask
process_mask_img is a subset of mask_img, it contains the voxels that should be processed (we only keep the slice z = 29 and the back of the brain to speed up computation)
import numpy as np
mask_img = load_img(haxby_dataset.mask)
# .astype() makes a copy.
process_mask = get_data(mask_img).astype(int)
picked_slice = 29
process_mask[..., (picked_slice + 1) :] = 0
process_mask[..., :picked_slice] = 0
process_mask[:, 30:] = 0
process_mask_img = new_img_like(mask_img, process_mask)
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_searchlight.py:61: UserWarning:
Data array used to create a new image contains 64-bit ints. This is likely due to creating the array with numpy and passing `int` as the `dtype`. Many tools such as FSL and SPM cannot deal with int64 in Nifti images, so for compatibility the data has been converted to int32.
Searchlight computation¶
Make processing parallel
Warning
As each thread will print its progress, n_jobs > 1 could mess up the information output.
n_jobs = 2
Define the cross-validation scheme used for validation. Here we use a KFold cross-validation on the run, which corresponds to splitting the samples in 4 folds and make 4 runs using each fold as a test set once and the others as learning sets
The radius is the one of the Searchlight sphere that will scan the volume
from sklearn.model_selection import KFold
from nilearn.decoding import SearchLight
cv = KFold(n_splits=4)
searchlight = SearchLight(
mask_img,
process_mask_img=process_mask_img,
radius=5.6,
n_jobs=n_jobs,
verbose=1,
cv=cv,
)
searchlight.fit(fmri_img, y)
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 2 out of 2 | elapsed: 13.4s finished
Visualization¶
%% After fitting the searchlight, we can access the searchlight scores as a NIfTI image using the scores_img_ attribute.
Use the fMRI mean image as a surrogate of anatomical data
Because scores are not a zero-center test statistics, we cannot use plot_stat_map
from nilearn.plotting import plot_img, plot_stat_map, show
plot_img(
scores_img,
bg_img=mean_fmri,
title="Searchlight scores image",
display_mode="z",
cut_coords=[-9],
vmin=0.2,
cmap="inferno",
threshold=0.2,
black_bg=True,
colorbar=True,
)
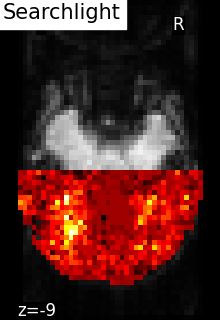
<nilearn.plotting.displays._slicers.ZSlicer object at 0x7f1ee8878ee0>
F-scores computation¶
from sklearn.feature_selection import f_classif
from nilearn.maskers import NiftiMasker
# For decoding, standardizing is often very important
nifti_masker = NiftiMasker(
mask_img=mask_img,
runs=run,
standardize="zscore_sample",
memory="nilearn_cache",
memory_level=1,
verbose=1,
)
fmri_masked = nifti_masker.fit_transform(fmri_img)
_, p_values = f_classif(fmri_masked, y)
p_values = -np.log10(p_values)
p_values[p_values > 10] = 10
p_unmasked = get_data(nifti_masker.inverse_transform(p_values))
# F_score results
p_ma = np.ma.array(p_unmasked, mask=np.logical_not(process_mask))
f_score_img = new_img_like(mean_fmri, p_ma)
plot_stat_map(
f_score_img,
mean_fmri,
title="F-scores",
display_mode="z",
cut_coords=[-9],
cmap="inferno",
)
show()
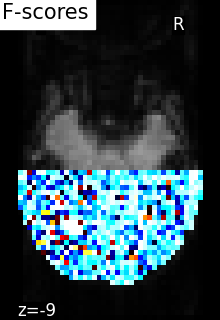
[NiftiMasker.wrapped] Loading mask from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c127a30>
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c126560>
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_searchlight.py:145: UserWarning:
[NiftiMasker.fit] Generation of a mask has been requested (imgs != None) while a mask was given at masker creation. Given mask will be used.
[NiftiMasker.wrapped] Resampling mask
[NiftiMasker.wrapped] Finished fit
________________________________________________________________________________
[Memory] Calling nilearn.maskers.nifti_masker.filter_and_mask...
filter_and_mask(<nibabel.nifti1.Nifti1Image object at 0x7f1f0c126560>, <nibabel.nifti1.Nifti1Image object at 0x7f1f0c8aa3b0>, { 'clean_args': None,
'clean_kwargs': {},
'cmap': 'gray',
'detrend': False,
'dtype': None,
'high_pass': None,
'high_variance_confounds': False,
'low_pass': None,
'reports': True,
'runs': 21 0
22 0
23 0
24 0
25 0
..
1398 11
1399 11
1400 11
1401 11
1402 11
Name: chunks, Length: 216, dtype: int64,
'smoothing_fwhm': None,
'standardize': 'zscore_sample',
'standardize_confounds': True,
't_r': None,
'target_affine': None,
'target_shape': None}, memory_level=1, memory=Memory(location=nilearn_cache/joblib), verbose=1, confounds=None, sample_mask=None, copy=True, dtype=None, sklearn_output_config=None)
[NiftiMasker.wrapped] Loading data from <nibabel.nifti1.Nifti1Image object at
0x7f1f0c126560>
[NiftiMasker.wrapped] Extracting region signals
[NiftiMasker.wrapped] Cleaning extracted signals
__________________________________________________filter_and_mask - 0.9s, 0.0min
[NiftiMasker.inverse_transform] Computing image from signals
________________________________________________________________________________
[Memory] Calling nilearn.masking.unmask...
unmask(array([5.229069, ..., 0.326513], shape=(39912,)), <nibabel.nifti1.Nifti1Image object at 0x7f1f0c8aa3b0>)
___________________________________________________________unmask - 0.1s, 0.0min
/home/runner/work/nilearn/nilearn/examples/02_decoding/plot_haxby_searchlight.py:164: UserWarning:
You are using the 'agg' matplotlib backend that is non-interactive.
No figure will be plotted when calling matplotlib.pyplot.show() or nilearn.plotting.show().
You can fix this by installing a different backend: for example via
pip install PyQt6
Total running time of the script: (0 minutes 24.877 seconds)
Estimated memory usage: 1013 MB