Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
nilearn.connectome.GroupSparseCovarianceCV¶
- class nilearn.connectome.GroupSparseCovarianceCV(alphas=4, n_refinements=4, cv=None, tol_cv=0.01, max_iter_cv=50, tol=0.001, max_iter=100, verbose=0, n_jobs=1, debug=False, early_stopping=True)[source]¶
Sparse inverse covariance w/ cross-validated choice of the parameter.
A cross-validated value for the regularization parameter is first determined using several calls to group_sparse_covariance. Then a final optimization is run to get a value for the precision matrices, using the selected value of the parameter. Different values of tolerance and of maximum iteration number can be used in these two phases (see the tol and tol_cv keyword below for example).
- Parameters:
- alphas
int, default=4 initial number of points in the grid of regularization parameter values. Each step of grid refinement adds that many points as well.
- n_refinements
int, default=4 number of times the initial grid should be refined.
- cv
int, default=None number of folds in a K-fold cross-validation scheme.
- tol_cv
float, default=1e-2 tolerance used to get the optimal alpha value. It has the same meaning as the tol parameter in
group_sparse_covariance.- max_iter_cv
int, default=50 maximum number of iterations for each optimization, during the alpha- selection phase.
- tol
float, default=1e-3 tolerance used during the final optimization for determining precision matrices value.
- max_iter
int, default=100 Maximum number of iterations for the solver.
- verbose
int, default=0 Verbosity level (0 means no message).
- n_jobs
int, default=1 The number of CPUs to use to do the computation. -1 means ‘all CPUs’.
- debug
bool, default=False if True, activates some internal checks for consistency. Only useful for nilearn developers, not users.
- early_stopping
bool, default=True if True, reduce computation time by using a heuristic to reduce the number of iterations required to get the optimal value for alpha. Be aware that this can lead to slightly different values for the optimal alpha compared to early_stopping=False.
- alphas
- Attributes:
- covariances_numpy.ndarray, shape (n_features, n_features, n_subjects)
covariance matrices, one per subject.
- precisions_numpy.ndarray, shape (n_features, n_features, n_subjects)
precision matrices, one per subject. All matrices have the same sparsity pattern (if a coefficient is zero for a given matrix, it is also zero for every other.)
- alpha_float
penalization parameter value selected.
- cv_alphas_list of floats
all values of the penalization parameter explored.
- cv_scores_numpy.ndarray, shape (n_alphas, n_folds)
scores obtained on test set for each value of the penalization parameter explored.
Notes
The search for the optimal penalization parameter (alpha) is done on an iteratively refined grid: first the cross-validated scores on a grid are computed, then a new refined grid is centered around the maximum, and so on.
- __init__(alphas=4, n_refinements=4, cv=None, tol_cv=0.01, max_iter_cv=50, tol=0.001, max_iter=100, verbose=0, n_jobs=1, debug=False, early_stopping=True)[source]¶
- fit(subjects, y=None)[source]¶
Compute cross-validated group-sparse precisions.
- Parameters:
- subjects
listof numpy.ndarray with shapes (n_samples, n_features) input subjects. Each subject is a 2D array, whose columns contain signals. Sample number can vary from subject to subject, but all subjects must have the same number of features (i.e. of columns.)
- yNone
This parameter is unused. It is solely included for scikit-learn compatibility.
- subjects
- Returns:
- selfGroupSparseCovarianceCV
the object instance itself.
- get_metadata_routing()¶
Get metadata routing of this object.
Please check User Guide on how the routing mechanism works.
- Returns:
- routingMetadataRequest
A
MetadataRequestencapsulating routing information.
- get_params(deep=True)¶
Get parameters for this estimator.
- Parameters:
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns:
- paramsdict
Parameter names mapped to their values.
- set_fit_request(*, subjects='$UNCHANGED$')¶
Request metadata passed to the
fitmethod.Note that this method is only relevant if
enable_metadata_routing=True(seesklearn.set_config). Please see User Guide on how the routing mechanism works.The options for each parameter are:
True: metadata is requested, and passed tofitif provided. The request is ignored if metadata is not provided.False: metadata is not requested and the meta-estimator will not pass it tofit.None: metadata is not requested, and the meta-estimator will raise an error if the user provides it.str: metadata should be passed to the meta-estimator with this given alias instead of the original name.
The default (
sklearn.utils.metadata_routing.UNCHANGED) retains the existing request. This allows you to change the request for some parameters and not others.Added in version 1.3.
Note
This method is only relevant if this estimator is used as a sub-estimator of a meta-estimator, e.g. used inside a
Pipeline. Otherwise it has no effect.- Parameters:
- subjectsstr, True, False, or None, default=sklearn.utils.metadata_routing.UNCHANGED
Metadata routing for
subjectsparameter infit.
- Returns:
- selfobject
The updated object.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters:
- **paramsdict
Estimator parameters.
- Returns:
- selfestimator instance
Estimator instance.
Examples using nilearn.connectome.GroupSparseCovarianceCV¶
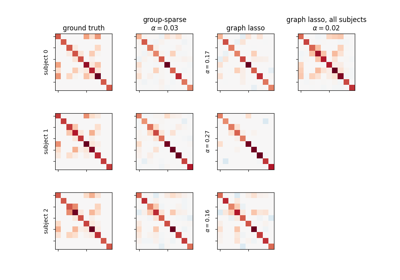
Connectivity structure estimation on simulated data
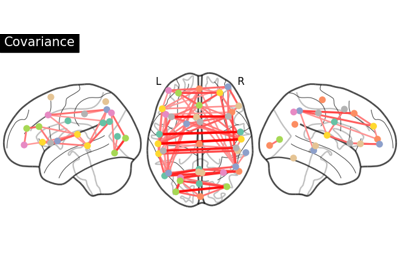
Group Sparse inverse covariance for multi-subject connectome