Note
This page is a reference documentation. It only explains the class signature, and not how to use it. Please refer to the user guide for the big picture.
8.8.7. nilearn.regions.RegionExtractor¶
- class nilearn.regions.RegionExtractor(maps_img, mask_img=None, min_region_size=1350, threshold=1.0, thresholding_strategy='ratio_n_voxels', extractor='local_regions', smoothing_fwhm=6, standardize=False, detrend=False, low_pass=None, high_pass=None, t_r=None, memory=Memory(location=None), memory_level=0, verbose=0)[source]¶
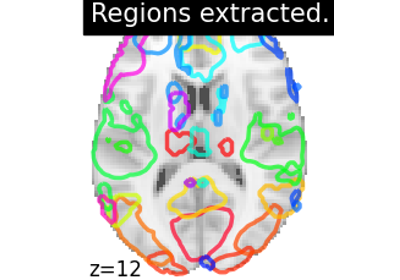
Class for brain region extraction.
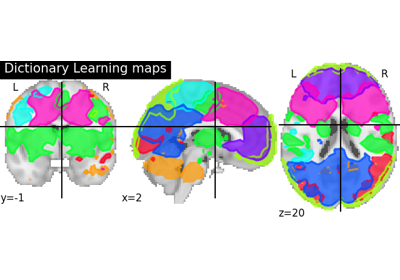
Region Extraction is a post processing technique which is implemented to automatically segment each brain atlas maps into different set of separated brain activated region. Particularly, to show that each decomposed brain maps can be used to focus on a target specific Regions of Interest analysis.
See [1].
New in version 0.2.
- Parameters
- maps_img4D Niimg-like object
Image containing a set of whole brain atlas maps or statistically decomposed brain maps.
- mask_imgNiimg-like object or None, optional
Mask to be applied to input data, passed to NiftiMapsMasker. If None, no masking is applied.
- min_region_sizefloat, optional
Minimum volume in mm3 for a region to be kept. For example, if the voxel size is 3x3x3 mm then the volume of the voxel is 27mm^3. Default=1350mm^3, which means we take minimum size of 1350 / 27 = 50 voxels.
- thresholdnumber, optional
A value used either in ratio_n_voxels or img_value or percentile thresholding_strategy based upon the choice of selection. Default=1.0.
- thresholding_strategystr {‘ratio_n_voxels’, ‘img_value’, ‘percentile’}, optional
If default ‘ratio_n_voxels’, we apply thresholding that will keep the more intense nonzero brain voxels (denoted as n_voxels) across all maps (n_voxels being the number of voxels in the brain volume). A float value given in threshold parameter indicates the ratio of voxels to keep meaning (if float=2. then maps will together have 2. x n_voxels non-zero voxels). If set to ‘percentile’, images are thresholded based on the score obtained with the given percentile on the data and the voxel intensities which are survived above this obtained score will be kept. If set to ‘img_value’, we apply thresholding based on the non-zero voxel intensities across all maps. A value given in threshold parameter indicates that we keep only those voxels which have intensities more than this value. Default=’ratio_n_voxels’.
- extractorstr {‘local_regions’, ‘connected_components’}, optional
If ‘connected_components’, each component/region in the image is extracted automatically by labelling each region based upon the presence of unique features in their respective regions. If ‘local_regions’, each component/region is extracted based on their maximum peak value to define a seed marker and then using random walker segementation algorithm on these markers for region separation. Default=’local_regions’.
- smoothing_fwhmscalar, optional
To smooth an image to extract most sparser regions. This parameter is passed to connected_regions and exists only for extractor ‘local_regions’. Please set this parameter according to maps resolution, otherwise extraction will fail. Default=6mm.
- standardizebool, optional
If True, the time series signals are centered and normalized by putting their mean to 0 and variance to 1. Recommended to set as True if signals are not already standardized. passed to class NiftiMapsMasker. Default=False.
- detrendbool, optional
This parameter is passed to nilearn.signal.clean basically indicates whether to detrend timeseries signals or not. passed to class NiftiMapsMasker. Default=False.
- low_passfloat, optional
This value will be applied on the signals by passing to signal.clean Please see the related documentation signal.clean for more details. passed to class NiftiMapsMasker.
- high_passfloat, optional
This value will be applied on the signals by passing to signal.clean Please see the related documentation signal.clean for more details. passed to NiftiMapsMasker.
- t_rfloat, optional
Repetition time in sec. This value is given to signal.clean Please see the related documentation for details. passed to NiftiMapsMasker.
- memoryinstance of joblib.Memory or string, optional
Used to cache the masking process. If a string is given, the path is set with this string as a folder name in the directory. passed to NiftiMapsMasker.
- memory_levelint, optional
Aggressiveness of memory catching. The higher the number, the higher the number of functions that will be cached. Zero mean no caching. passed to NiftiMapsMasker. Default=0.
- verboseint, optional
Indicates the level of verbosity by printing the message. Zero indicates nothing is printed. Default=0.
See also
nilearn.regions.connected_label_regionsA function can be readily used for extraction of regions on labels based atlas images.
References
- 1
Abraham et al. “Region segmentation for sparse decompositions: better brain parcellations from rest fMRI”, Sparsity Techniques in Medical Imaging, Sep 2014, Boston, United States. pp.8
- Attributes
- `index_`numpy array
Array of list of indices where each index value is assigned to each separate region of its corresponding family of brain maps.
- `regions_img_`Nifti1Image
List of separated regions with each region lying on an original volume concatenated into a 4D image.
- __init__(maps_img, mask_img=None, min_region_size=1350, threshold=1.0, thresholding_strategy='ratio_n_voxels', extractor='local_regions', smoothing_fwhm=6, standardize=False, detrend=False, low_pass=None, high_pass=None, t_r=None, memory=Memory(location=None), memory_level=0, verbose=0)[source]¶
Initialize self. See help(type(self)) for accurate signature.
- fit_transform(imgs, confounds=None, sample_mask=None)[source]¶
Prepare and perform signal extraction.
- get_params(deep=True)¶
Get parameters for this estimator.
- Parameters
- deepbool, default=True
If True, will return the parameters for this estimator and contained subobjects that are estimators.
- Returns
- paramsdict
Parameter names mapped to their values.
- inverse_transform(region_signals)[source]¶
Compute voxel signals from region signals
Any mask given at initialization is taken into account.
- Parameters
- region_signals2D numpy.ndarray
Signal for each region. shape: (number of scans, number of regions)
- Returns
- voxel_signalsnibabel.Nifti1Image
Signal for each voxel. shape: that of maps.
- set_params(**params)¶
Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as
Pipeline). The latter have parameters of the form<component>__<parameter>so that it’s possible to update each component of a nested object.- Parameters
- **paramsdict
Estimator parameters.
- Returns
- selfestimator instance
Estimator instance.
- transform(imgs, confounds=None, sample_mask=None)[source]¶
Apply mask, spatial and temporal preprocessing
- Parameters
- imgs3D/4D Niimg-like object
See http://nilearn.github.io/manipulating_images/input_output.html Images to process. It must boil down to a 4D image with scans number as last dimension.
- confoundsCSV file or array-like, optional
This parameter is passed to signal.clean. Please see the related documentation for details. shape: (number of scans, number of confounds)
- sample_maskAny type compatible with numpy-array indexing, optional
shape: (number of scans - number of volumes removed, ) Masks the niimgs along time/fourth dimension to perform scrubbing (remove volumes with high motion) and/or non-steady-state volumes. This parameter is passed to signal.clean.
New in version 0.8.0.
- Returns
- region_signals2D numpy.ndarray
Signal for each element. shape: (number of scans, number of elements)
- transform_single_imgs(imgs, confounds=None, sample_mask=None)[source]¶
Extract signals from a single 4D niimg.
- Parameters
- imgs3D/4D Niimg-like object
See http://nilearn.github.io/manipulating_images/input_output.html Images to process. It must boil down to a 4D image with scans number as last dimension.
- confoundsCSV file or array-like, optional
This parameter is passed to signal.clean. Please see the related documentation for details. shape: (number of scans, number of confounds)
- sample_maskAny type compatible with numpy-array indexing, optional
shape: (number of scans - number of volumes removed, ) Masks the niimgs along time/fourth dimension to perform scrubbing (remove volumes with high motion) and/or non-steady-state volumes. This parameter is passed to signal.clean.
New in version 0.8.0.
- Returns
- region_signals2D numpy.ndarray
Signal for each map. shape: (number of scans, number of maps)